Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines
- PMID: 16789826
- PMCID: PMC1523251
- DOI: 10.1371/journal.pgen.0020120
Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines
Abstract
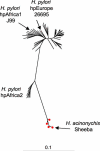
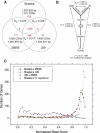
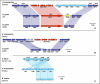
Helicobacter pylori infection of humans is so old that its population genetic structure reflects that of ancient human migrations. A closely related species, Helicobacter acinonychis, is specific for large felines, including cheetahs, lions, and tigers, whereas hosts more closely related to humans harbor more distantly related Helicobacter species. This observation suggests a jump between host species. But who ate whom and when did it happen? In order to resolve this question, we determined the genomic sequence of H. acinonychis strain Sheeba and compared it to genomes from H. pylori. The conserved core genes between the genomes are so similar that the host jump probably occurred within the last 200,000 (range 50,000-400,000) years. However, the Sheeba genome also possesses unique features that indicate the direction of the host jump, namely from early humans to cats. Sheeba possesses an unusually large number of highly fragmented genes, many encoding outer membrane proteins, which may have been destroyed in order to bypass deleterious responses from the feline host immune system. In addition, the few Sheeba-specific genes that were found include a cluster of genes encoding sialylation of the bacterial cell surface carbohydrates, which were imported by horizontal genetic exchange and might also help to evade host immune defenses. These results provide a genomic basis for elucidating molecular events that allow bacteria to adapt to novel animal hosts.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

