Retroviral DNA integration: viral and cellular determinants of target-site selection
- PMID: 16789841
- PMCID: PMC1480600
- DOI: 10.1371/journal.ppat.0020060
Retroviral DNA integration: viral and cellular determinants of target-site selection
Abstract
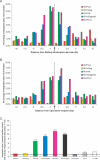
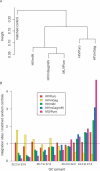
Retroviruses differ in their preferences for sites for viral DNA integration in the chromosomes of infected cells. Human immunodeficiency virus (HIV) integrates preferentially within active transcription units, whereas murine leukemia virus (MLV) integrates preferentially near transcription start sites and CpG islands. We investigated the viral determinants of integration-site selection using HIV chimeras with MLV genes substituted for their HIV counterparts. We found that transferring the MLV integrase (IN) coding region into HIV (to make HIVmIN) caused the hybrid to integrate with a specificity close to that of MLV. Addition of MLV gag (to make HIVmGagmIN) further increased the similarity of target-site selection to that of MLV. A chimeric virus with MLV Gag only (HIVmGag) displayed targeting preferences different from that of both HIV and MLV, further implicating Gag proteins in targeting as well as IN. We also report a genome-wide analysis indicating that MLV, but not HIV, favors integration near DNase I-hypersensitive sites (i.e., +/- 1 kb), and that HIVmIN and HIVmGagmIN also favored integration near these features. These findings reveal that IN is the principal viral determinant of integration specificity; they also reveal a new role for Gag-derived proteins, and strengthen models for integration targeting based on tethering of viral IN proteins to host proteins.
Conflict of interest statement
Figures



References
-
- Check E. Gene therapy put on hold as third child develops cancer. Nature. 2005;433:561. - PubMed
-
- Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. - PubMed
-
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

