Intrastrain heterogeneity of the mgpB gene in Mycoplasma genitalium is extensive in vitro and in vivo and suggests that variation is generated via recombination with repetitive chromosomal sequences
- PMID: 16790744
- PMCID: PMC1489687
- DOI: 10.1128/IAI.00239-06
Intrastrain heterogeneity of the mgpB gene in Mycoplasma genitalium is extensive in vitro and in vivo and suggests that variation is generated via recombination with repetitive chromosomal sequences
Abstract
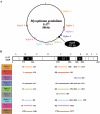
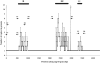
Mycoplasma genitalium is associated with reproductive tract disease in women and may persist in the lower genital tract for months, potentially increasing the risk of upper tract infection and transmission to uninfected partners. Despite its exceptionally small genome (580 kb), approximately 4% is composed of repeated elements known as MgPar sequences (MgPa repeats) based on their homology to the mgpB gene that encodes the immunodominant MgPa adhesin protein. The presence of these MgPar sequences, as well as mgpB variability between M. genitalium strains, suggests that mgpB and MgPar sequences recombine to produce variant MgPa proteins. To examine the extent and generation of diversity within single strains of the organism, we examined mgpB variation within M. genitalium strain G-37 and observed sequence heterogeneity that could be explained by recombination between the mgpB expression site and putative donor MgPar sequences. Similarly, we analyzed mgpB sequences from cervical specimens from a persistently infected woman (21 months) and identified 17 different mgpB variants within a single infecting M. genitalium strain, confirming that mgpB heterogeneity occurs over the course of a natural infection. These observations support the hypothesis that recombination occurs between the mgpB gene and MgPar sequences and that the resulting antigenically distinct MgPa variants may contribute to immune evasion and persistence of infection.
Figures



References
-
- Amundsen, S. K., and G. R. Smith. 2003. Interchangeable parts of the Escherichia coli recombination machinery. Cell 112:741-744. - PubMed
-
- Aravind, L., and E. V. Koonin. 1998. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem. Sci. 23:17-19. - PubMed
-
- Bjornelius, E., P. Lidbrink, and J. S. Jensen. 2000. Mycoplasma genitalium in non-gonococcal urethritis—a study in Swedish male STD patients. Int. J. STD AIDS 11:292-296. - PubMed
-
- Carvalho, F. M., M. M. Fonseca, S. Batistuzzo De Medeiros, K. C. Scortecci, C. A. Blaha, and L. F. Agnez-Lima. 2005. DNA repair in reduced genome: the mycoplasma model. Gene 360:111-119. - PubMed
-
- Centurion-Lara, A., R. E. LaFond, K. Hevner, C. Godornes, B. J. Molini, W. C. Van Voorhis, and S. A. Lukehart. 2004. Gene conversion: a mechanism for generation of heterogeneity in the tprK gene of Treponema pallidum during infection. Mol. Microbiol. 52:1579-1596. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

