Efficient amplification and cloning of near full-length hepatitis C virus genome from clinical samples
- PMID: 16793008
- PMCID: PMC7092855
- DOI: 10.1016/j.bbrc.2006.06.039
Efficient amplification and cloning of near full-length hepatitis C virus genome from clinical samples
Abstract
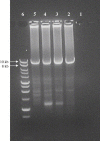
Long RT-PCR (LRP) amplification of RNA templates is sometimes difficult compared to long PCR of DNA templates. Among RNA templates, hepatitis C virus (HCV) represents an excellent example to challenge the potential of LRP technology due to its extensive secondary structures and its difficulty to be readily cultured in vitro. The only source for viral genome amplification is clinical samples in which HCV is usually present at low titers. We have created a comprehensive optimization protocol that allows robust amplification of a 9.1 kb fragment of HCV, followed by efficient cloning into a novel vector. Detailed analyses indicate the lack of potential LRP-mediated recombination and the preservation of viral diversity. Thus, our LRP protocol could be applied for the amplification of other difficult RNA templates and may facilitate RNA virus research such as linked viral mutations and reverse genetics.
Figures


Similar articles
-
RT-PCR amplification and cloning of large viral sequences.Methods Mol Biol. 2010;630:139-49. doi: 10.1007/978-1-60761-629-0_10. Methods Mol Biol. 2010. PMID: 20300996
-
Separation of near full-length hepatitis C virus quasispecies variants from a complex population.J Virol Methods. 2007 May;141(2):220-4. doi: 10.1016/j.jviromet.2006.12.002. Epub 2007 Jan 8. J Virol Methods. 2007. PMID: 17208310
-
Full-Length Open Reading Frame Amplification of Hepatitis C Virus.Methods Mol Biol. 2019;1911:85-91. doi: 10.1007/978-1-4939-8976-8_5. Methods Mol Biol. 2019. PMID: 30593619
-
[Heterogeneity of hepatitis C virus (HCV) genomes and primer selection for detecting of HCV virus RNA with polymerase chain reaction assay].Nihon Rinsho. 1992 Jul;50 Suppl:279-84. Nihon Rinsho. 1992. PMID: 1328721 Review. Japanese. No abstract available.
-
Detection of hepatitis C virus RNA: application to diagnostics and research.Curr Stud Hematol Blood Transfus. 1994;(61):49-68. doi: 10.1159/000423268. Curr Stud Hematol Blood Transfus. 1994. PMID: 7956330 Review. No abstract available.
Cited by
-
Comprehensive cloning of patient-derived 9022-bp amplicons of hepatitis C virus.J Virol Methods. 2013 Aug;191(2):105-12. doi: 10.1016/j.jviromet.2013.04.010. Epub 2013 Apr 17. J Virol Methods. 2013. PMID: 23602804 Free PMC article.
-
Development of a sensitive RT-PCR method for amplifying and sequencing near full-length HCV genotype 1 RNA from patient samples.Virol J. 2013 Feb 12;10:53. doi: 10.1186/1743-422X-10-53. Virol J. 2013. PMID: 23402332 Free PMC article.
-
Viral categorization and discovery in human circulation by transcriptome sequencing.Biochem Biophys Res Commun. 2013 Jul 5;436(3):525-9. doi: 10.1016/j.bbrc.2013.05.139. Epub 2013 Jun 11. Biochem Biophys Res Commun. 2013. PMID: 23764402 Free PMC article.
-
Hepatitis C virus diversity and evolution in the full open-reading frame during antiviral therapy.PLoS One. 2008 May 7;3(5):e2123. doi: 10.1371/journal.pone.0002123. PLoS One. 2008. PMID: 18463735 Free PMC article.
-
Comparative analysis of nearly full-length hepatitis C virus quasispecies from patients experiencing viral breakthrough during antiviral therapy: clustered mutations in three functional genes, E2, NS2, and NS5a.J Virol. 2008 Oct;82(19):9417-24. doi: 10.1128/JVI.00896-08. Epub 2008 Jul 30. J Virol. 2008. PMID: 18667493 Free PMC article.
References
-
- Chumakov K.M. Reverse transcriptase can inhibit PCR and stimulate primer-dimer formation. PCR Meth. Appl. 1994;4:62–64. - PubMed
-
- Tuplin A., Evans D.J., Simmonds P. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J. Gen. Virol. 2004;85:3037–3047. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

