Os8N3 is a host disease-susceptibility gene for bacterial blight of rice
- PMID: 16798873
- PMCID: PMC1502487
- DOI: 10.1073/pnas.0604088103
Os8N3 is a host disease-susceptibility gene for bacterial blight of rice
Abstract
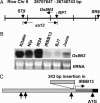
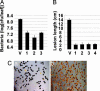
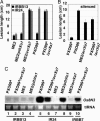
Many bacterial diseases of plants depend on the interaction of type III effector genes of the pathogen and disease-susceptibility genes of the host. The host susceptibility genes are largely unknown. Here, we show that expression of the rice gene Os8N3, a member of the MtN3 gene family from plants and animals, is elevated upon infection by Xanthomonas oryzae pv. oryzae strain PXO99(A) and depends on the type III effector gene pthXo1. Os8N3 resides near xa13, and PXO99(A) failed to induce Os8N3 in rice lines with xa13. Silencing of Os8N3 by inhibitory RNA produced plants that were resistant to infection by strain PXO99(A) yet remained susceptible to other strains of the pathogen. The effector gene avrXa7 from strain PXO86 enabled PXO99(A) compatibility on either xa13- or Os8N3-silenced plants. The findings indicate that Os8N3 is a host susceptibility gene for bacterial blight targeted by the type III effector PthXo1. The results support the hypothesis that X. oryzae pv. oryzae commandeers the regulation of otherwise developmentally regulated host genes to induce a state of disease susceptibility. Furthermore, the results support a model in which the pathogen induces disease susceptibility in a gene-for-gene manner.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures




Similar articles
-
Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAgamma1 and OsTFX1 during bacterial blight of rice.Proc Natl Acad Sci U S A. 2007 Jun 19;104(25):10720-5. doi: 10.1073/pnas.0701742104. Epub 2007 Jun 11. Proc Natl Acad Sci U S A. 2007. PMID: 17563377 Free PMC article.
-
Designer TAL effectors induce disease susceptibility and resistance to Xanthomonas oryzae pv. oryzae in rice.Mol Plant. 2013 May;6(3):781-9. doi: 10.1093/mp/sst034. Epub 2013 Feb 21. Mol Plant. 2013. PMID: 23430045
-
Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3.Plant Cell. 2010 Nov;22(11):3864-76. doi: 10.1105/tpc.110.078964. Epub 2010 Nov 23. Plant Cell. 2010. PMID: 21098734 Free PMC article.
-
Recessive resistance genes and the Oryza sativa-Xanthomonas oryzae pv. oryzae pathosystem.Mol Plant Microbe Interact. 2007 Jul;20(7):731-9. doi: 10.1094/MPMI-20-7-0731. Mol Plant Microbe Interact. 2007. PMID: 17601161 Review.
-
Host and pathogen factors controlling the rice-Xanthomonas oryzae interaction.Plant Physiol. 2009 Aug;150(4):1677-86. doi: 10.1104/pp.109.139360. Epub 2009 May 20. Plant Physiol. 2009. PMID: 19458115 Free PMC article. Review. No abstract available.
Cited by
-
Xa7, a new executor R gene that confers durable and broad-spectrum resistance to bacterial blight disease in rice.Plant Commun. 2021 Jan 9;2(3):100143. doi: 10.1016/j.xplc.2021.100143. eCollection 2021 May 10. Plant Commun. 2021. PMID: 34027390 Free PMC article.
-
Suppression of Xo1-Mediated Disease Resistance in Rice by a Truncated, Non-DNA-Binding TAL Effector of Xanthomonas oryzae.Front Plant Sci. 2016 Oct 13;7:1516. doi: 10.3389/fpls.2016.01516. eCollection 2016. Front Plant Sci. 2016. PMID: 27790231 Free PMC article.
-
From perception to activation: the molecular-genetic and biochemical landscape of disease resistance signaling in plants.Arabidopsis Book. 2010;8:e012. doi: 10.1199/tab.0124. Epub 2010 May 14. Arabidopsis Book. 2010. PMID: 22303251 Free PMC article.
-
Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis.BMC Genomics. 2015 Jul 11;16(1):520. doi: 10.1186/s12864-015-1730-y. BMC Genomics. 2015. PMID: 26162601 Free PMC article.
-
Common bean resistance to Xanthomonas is associated with upregulation of the salicylic acid pathway and downregulation of photosynthesis.BMC Genomics. 2020 Aug 18;21(1):566. doi: 10.1186/s12864-020-06972-6. BMC Genomics. 2020. PMID: 32811445 Free PMC article.
References
-
- Belkhadir Y., Subramaniam R., Dangl J. L. Curr. Opin. Plant Biol. 2004;7:391–399. - PubMed
-
- He S. Y., Nomura K., Whittam T. S. Biochim. Biophys. Acta. 2004;1694:181–206. - PubMed
-
- Mota L. J., Cornelis G. R. Ann. Med. 2005;37:234–249. - PubMed
-
- Mudgett M. B. Annu. Rev. Plant Biol. 2005;56:509–531. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

