MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis
- PMID: 16798886
- PMCID: PMC1488920
- DOI: 10.1105/tpc.106.042127
MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis
Abstract
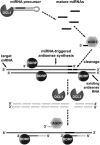
ARGONAUTE1 (AGO1) of Arabidopsis thaliana mediates the cleavage of microRNA (miRNA)-targeted mRNAs, and it has also been implicated in the posttranscriptional silencing of transgenes and the maintenance of chromatin structure. Mutations in AGO1 severely disrupt plant development, indicating that miRNA function and possibly other aspects of RNA interference are essential for maintaining normal patterns of gene expression. Using microarrays, we found that 1 to 6% of genes display significant expression changes in several alleles of ago1 at multiple developmental stages, with the majority showing higher levels. Several classes of known miRNA targets increased markedly in ago1, whereas others showed little or no change. Cleavage of mRNAs within miRNA-homologous sites was reduced but not abolished in an ago1 -null background, indicating that redundant slicer activity exists in Arabidopsis. Small interfering RNAs and larger 30- to 60-nucleotide RNA fragments corresponding to highly upregulated miRNA target genes accumulated in wild-type plants but not in ago1, the RNA-dependent RNA polymerase mutants rdr2 and rdr6, or the Dicer-like mutants dcl1 and dcl3. Both sense and antisense RNAs corresponding to these miRNA targets accumulated in the ago1 and dcl1 backgrounds. These results indicate that a subset of endogenous mRNA targets of RNA interference may be regulated through a mechanism of second-strand RNA synthesis and degradation initiated by or in addition to miRNA-mediated cleavage.
Figures






References
-
- Allen, E., Xie, Z., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. - PubMed
-
- Bao, N., Lye, K.W., and Barton, M.K. (2004). MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 7 653–662. - PubMed
-
- Baulcombe, D. (2005). RNA silencing. Trends Biochem. Sci. 30 290–293. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

