Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes
- PMID: 16798940
- PMCID: PMC1533958
- DOI: 10.1104/pp.106.082826
Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes
Abstract
The Dof (DNA binding with one finger) transcriptional activator rice (Oryza sativa) prolamin box binding factor (RPBF), which is involved in gene regulation of rice seed storage proteins, has been isolated from rice cDNA expressed sequence tag clones containing the conserved Dof. RPBF is found as a single gene per haploid genome. Comparison of RPBF genomic and cDNA sequences revealed that the genomic copy is interrupted by one long intron of 1,892 bp in the 5' noncoding region. We demonstrated by transient expression in rice callus protoplasts that the isolated RPBF trans-activated several storage protein genes via an AAAG target sequence located within their promoters, and with methylation interference experiments the additional AAAG-like sequences in promoters of genes expressed in maturing seeds were recognized by the RPBF protein. Binding was sequence specific, since mutation of the AAAG motif or its derivatives decreased both binding and trans-activation by RPBF. Synergism between RPBF and RISBZ1 recognizing the GCN4 motif [TGA(G/C)TCA] was observed in the expression of many storage protein genes. Overexpression of both transcription factors gave rise to much higher levels of expression than the sum of individual activities elicited by either RPBF or RISBZ1 alone. Furthermore, mutation of recognition sites suppressed reciprocal trans-activation ability, indicating that there are mutual interactions between RISBZ1 and RPBF. The RPBF gene is predominantly expressed in maturing endosperm and coordinately expressed with seed storage protein genes, and is involved in the quantitative regulation of genes expressed in the endosperm in cooperation with RISBZ1.
Figures




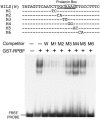


References
-
- Adachi T, Izumi H, Yamada T, Tanaka K, Takeuchi S, Nakamura R, Matsuda T (1993) Gene structure and expression of rice seed allergenic proteins belonging to the alpha-amylase/trypsin inhibitor family. Plant Mol Biol 21: 239–248 - PubMed
-
- Conlan RS, Hammond-Kosack M, Bevan M (1999) Transcription activation mediated by the bZIP factor SPA on the endosperm box is modulated by ESBF-1 in vitro. Plant J 19: 173–181 - PubMed
-
- Diaz I, Vicente-Carbajosa J, Abraham Z, Martinez M, Isabel-LaMoneda I, Carbonero P (2002) The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J 29: 453–464 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

