Natural variation for carbohydrate content in Arabidopsis. Interaction with complex traits dissected by quantitative genetics
- PMID: 16798941
- PMCID: PMC1533913
- DOI: 10.1104/pp.106.082396
Natural variation for carbohydrate content in Arabidopsis. Interaction with complex traits dissected by quantitative genetics
Abstract
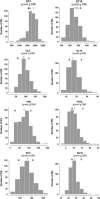
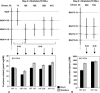
Besides being a metabolic fuel, carbohydrates play important roles in plant growth and development, in stress responses, and as signal molecules. We exploited natural variation in Arabidopsis (Arabidopsis thaliana) to decipher the genetic architecture determining carbohydrate content. A quantitative trait locus (QTL) approach in the Bay-0 x Shahdara progeny grown in two contrasting nitrogen environments led to the identification of 39 QTLs for starch, glucose, fructose, and sucrose contents representing at least 14 distinct polymorphic loci. A major QTL for fructose content (FR3.4) and a QTL for starch content (ST3.4) were confirmed in heterogeneous inbred families. Several genes associated with carbon (C) metabolism colocalize with the identified QTL. QTLs for senescence-related traits, and for flowering time, water status, and nitrogen-related traits, previously detected with the same genetic material, colocalize with C-related QTLs. These colocalizations reflect the complex interactions of C metabolism with other physiological processes. QTL fine-mapping and cloning could thus lead soon to the identification of genes potentially involved in the control of different connected physiological processes.
Figures
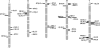
References
-
- Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5: 22–29 - PubMed
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

