Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase
- PMID: 16798944
- PMCID: PMC1533943
- DOI: 10.1104/pp.106.082198
Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase
Abstract
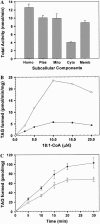
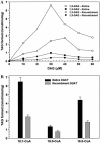
Triacylglycerols (TAGs) are the most important storage form of energy for eukaryotic cells. TAG biosynthetic activity was identified in the cytosolic fraction of developing peanut (Arachis hypogaea) cotyledons. This activity was NaF insensitive and acyl-coenzyme A (CoA) dependent. Acyl-CoA:diacylglycerol acyltransferase (DGAT) catalyzes the final step in TAG biosynthesis that acylates diacylglycerol to TAG. Soluble DGAT was identified from immature peanuts and purified by conventional column chromatographic procedures. The enzyme has a molecular mass of 41 +/- 1.0 kD. Based on the partial peptide sequence, a degenerate probe was used to obtain the full-length cDNA. The isolated gene shared less than 10% identity with the previously identified DGAT1 and 2 families, but has 13% identity with the bacterial bifunctional wax ester/DGAT. To differentiate the unrelated families, we designate the peanut gene as AhDGAT. Expression of peanut cDNA in Escherichia coli resulted in the formation of labeled TAG and wax ester from [14C]acetate. The recombinant E. coli showed high levels of DGAT activity but no wax ester synthase activity. TAGs were localized in transformed cells with Nile blue A and oil red O staining. The recombinant and native DGAT was specific for 1,2-diacylglycerol and did not utilize hexadecanol, glycerol-3-phosphate, monoacylglycerol, lysophosphatidic acid, and lysophosphatidylcholine. Oleoyl-CoA was the preferred acyl donor as compared to palmitoyl- and stearoyl-CoAs. These data suggest that the cytosol is one of the sites for TAG biosynthesis in oilseeds. The identified pathway may present opportunities of bioengineering oil-yielding plants for increased oil production.
Figures






References
-
- Bouvier-Nave P, Benveniste P, Oelkers P, Sturley SL, Schaller H (2000) Expression in yeast and tobacco of plant cDNAs encoding acyl CoA:diacylglycerol acyltransferase. Eur J Biochem 267: 85–96 - PubMed
-
- Broun P, Gettner S, Somerville C (1999) Genetic engineering of plant lipids. Annu Rev Nutr 19: 197–216 - PubMed
-
- Daniel J, Deb C, Dubey VS, Sirakova TD, Abomoelak B, Morbidoni HR, Kolattukudy PE (2004) Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol 186: 5017–5030 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

