Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles
- PMID: 16800888
- PMCID: PMC1525188
- DOI: 10.1186/1471-2164-7-162
Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles
Abstract
Background: Pseudomonas aeruginosa is a genetically complex bacterium which can adopt and switch between a free-living or biofilm lifestyle, a versatility that enables it to thrive in many different environments and contributes to its success as a human pathogen.
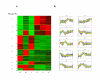
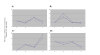
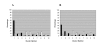
Results: Transcriptomes derived from growth states relevant to the lifestyle of P. aeruginosa were clustered using three different methods (K-means, K-means spectral and hierarchical clustering). The culture conditions used for this study were; biofilms incubated for 8, 14, 24 and 48 hrs, and planktonic culture (logarithmic and stationary phase). This cluster analysis revealed the existence and provided a clear illustration of distinct expression profiles present in the dataset. Moreover, it gave an insight into which genes are up-regulated in planktonic, developing biofilm and confluent biofilm states. In addition, this analysis confirmed the contribution of quorum sensing (QS) and RpoS regulated genes to the biofilm mode of growth, and enabled the identification of a 60.69 Kbp region of the genome associated with stationary phase growth (stationary phase planktonic culture and confluent biofilms).
Conclusion: This is the first study to use clustering to separate a large P. aeruginosa microarray dataset consisting of transcriptomes obtained from diverse conditions relevant to its growth, into different expression profiles. These distinct expression profiles not only reveal novel aspects of P. aeruginosa gene expression but also provide a growth specific transcriptomic reference dataset for the research community.
Figures
References
-
- Bodey GP, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5:279–313. - PubMed
-
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

