Wild-type and central DNA flap defective HIV-1 lentiviral vector genomes: intracellular visualization at ultrastructural resolution levels
- PMID: 16800894
- PMCID: PMC1538615
- DOI: 10.1186/1742-4690-3-38
Wild-type and central DNA flap defective HIV-1 lentiviral vector genomes: intracellular visualization at ultrastructural resolution levels
Abstract
HIV-1 and other lentiviruses have the unique ability among retroviruses to efficiently replicate in non-dividing cells as a result of the active nuclear import of their DNA genome across an interphasic nuclear membrane. Previous work has shown that a three-stranded DNA structure synthesized during HIV-1 reverse transcription, called the central DNA flap, acts as a cis-determinant of HIV-1 genome nuclear import. Concordantly, DNA Flap re-insertion in lentiviral-derived gene therapy vectors stimulates gene transfer efficiencies and complements the level of nuclear import to wild-type levels quantitatively indistinguishable from wild-type virus in all cell types and tissues examined so far. In order to define the precise nature of the replicative defect of DNA flap mutant viruses, we carried out in situ DNA hybridization experiments with electron microscopy to determine the subcellular localization of DNA flap mutant and wild-type HIV-1 genomes. We found that Flap defective DNA genomes accumulate at the cytoplasmic face of the nuclear membrane with no overlap across the nuclear membrane, whereas wild-type genomes localize throughout the nuclear compartment. These data provide an unequivocal confirmation of the role of the DNA flap in HIV-1 nuclear import and further establish that the DNA flap controls a step that immediately precedes translocation through the nuclear pore. Further, the widespread distribution of wild-type genomes within the open chromatin confirms the recent genome-wide mapping of HIV-1 cDNA integration sites and points to an as-yet poorly understood step of intranuclear transport of HIV-1 pre-integration complexes.
Figures
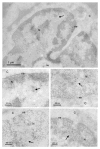
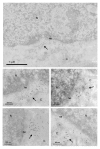

References
-
- Sirven A, Pflumio F, Zennou V, Titeux M, Vainchenker W, Coulombel L, Dubart-Kupperschmitt A, Charneau P. The human immunodeficiency virus type-1 central DNA Flap is a crucial determinant for lentiviral vector nuclear import and gene transduction of human hematopoietic stem cells. Blood. 2000;96:4103–4110. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

