Identification of putative domain linkers by a neural network - application to a large sequence database
- PMID: 16800897
- PMCID: PMC1538634
- DOI: 10.1186/1471-2105-7-323
Identification of putative domain linkers by a neural network - application to a large sequence database
Abstract
Background: The reliable dissection of large proteins into structural domains represents an important issue for structural genomics/proteomics projects. To provide a practical approach to this issue, we tested the ability of neural network to identify domain linkers from the SWISSPROT database (101602 sequences).
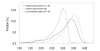
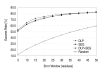
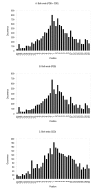
Results: Our search detected 3009 putative domain linkers adjacent to or overlapping with domains, as defined by sequence similarity to either Protein Data Bank (PDB) or Conserved Domain Database (CDD) sequences. Among these putative linkers, 75% were "correctly" located within 20 residues of a domain terminus, and the remaining 25% were found in the middle of a domain, and probably represented failed predictions. Moreover, our neural network predicted 5124 putative domain linkers in structurally un-annotated regions without sequence similarity to PDB or CDD sequences, which suggest to the possible existence of novel structural domains. As a comparison, we performed the same analysis by identifying low-complexity regions (LCR), which are known to encode unstructured polypeptide segments, and observed that the fraction of LCRs that correlate with domain termini is similar to that of domain linkers. However, domain linkers and LCRs appeared to identify different types of domain boundary regions, as only 32% of the putative domain linkers overlapped with LCRs.
Conclusion: Overall, our study indicates that the two methods detect independent and complementary regions, and that the combination of these methods can substantially improve the sensitivity of the domain boundary prediction. This finding should enable the identification of novel structural domains, yielding new targets for large scale protein analyses.
Figures

References
-
- Mallick P, Goodwill KE, Fitz-Gibbon S, Miller JH, Eisenberg D. Selecting protein targets for structural genomics of Pyrobaculum aerophilum: validating automated fold assignment methods by using binary hypothesis testing. Proc Natl Acad Sci U S A. 2000;97:2450–2455. doi: 10.1073/pnas.050589297. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

