A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors
- PMID: 16801547
- PMCID: PMC1482798
- DOI: 10.1073/pnas.0601456103
A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors
Erratum in
- Proc Natl Acad Sci U S A. 2006 Oct 31;103(44):16614
Abstract
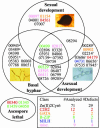
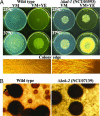
The low rate of homologous recombination exhibited by wild-type strains of filamentous fungi has hindered development of high-throughput gene knockout procedures for this group of organisms. In this study, we describe a method for rapidly creating knockout mutants in which we make use of yeast recombinational cloning, Neurospora mutant strains deficient in nonhomologous end-joining DNA repair, custom-written software tools, and robotics. To illustrate our approach, we have created strains bearing deletions of 103 Neurospora genes encoding transcription factors. Characterization of strains during growth and both asexual and sexual development revealed phenotypes for 43% of the deletion mutants, with more than half of these strains possessing multiple defects. Overall, the methodology, which achieves high-throughput gene disruption at an efficiency >90% in this filamentous fungus, promises to be applicable to other eukaryotic organisms that have a low frequency of homologous recombination.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

