Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids
- PMID: 16801548
- PMCID: PMC1502431
- DOI: 10.1073/pnas.0601167103
Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids
Abstract
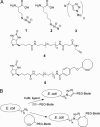
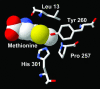
The incorporation of noncanonical amino acids into recombinant proteins in Escherichia coli can be facilitated by the introduction of new aminoacyl-tRNA synthetase activity into the expression host. We describe here a screening procedure for the identification of new aminoacyl-tRNA synthetase activity based on the cell surface display of noncanonical amino acids. Screening of a saturation mutagenesis library of the E. coli methionyl-tRNA synthetase (MetRS) led to the discovery of three MetRS mutants capable of incorporating the long-chain amino acid azidonorleucine into recombinant proteins with modest efficiency. The Leu-13 --> Gly (L13G) mutation is found in each of the three MetRS mutants, and the MetRS variant containing this single mutation is highly efficient in producing recombinant proteins that contain azidonorleucine.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



Similar articles
-
Discovery of Escherichia coli methionyl-tRNA synthetase mutants for efficient labeling of proteins with azidonorleucine in vivo.Proc Natl Acad Sci U S A. 2009 Sep 8;106(36):15285-90. doi: 10.1073/pnas.0905735106. Epub 2009 Aug 17. Proc Natl Acad Sci U S A. 2009. PMID: 19706454 Free PMC article.
-
Directed Evolution of Orthogonal Pyrrolysyl-tRNA Synthetases in Escherichia coli for the Genetic Encoding of Noncanonical Amino Acids.Methods Mol Biol. 2018;1728:97-111. doi: 10.1007/978-1-4939-7574-7_5. Methods Mol Biol. 2018. PMID: 29404992
-
Effect of a domain-spanning disulfide on aminoacyl-tRNA synthetase activity.Biochemistry. 2009 Oct 27;48(42):10113-9. doi: 10.1021/bi9012275. Biochemistry. 2009. PMID: 19772352
-
Substrate selection by aminoacyl-tRNA synthetases.Nucleic Acids Symp Ser. 1995;(33):40-2. Nucleic Acids Symp Ser. 1995. PMID: 8643392 Review.
-
Non-canonical Amino Acid Substrates of E. coli Aminoacyl-tRNA Synthetases.Chembiochem. 2022 Jan 5;23(1):e202100299. doi: 10.1002/cbic.202100299. Epub 2021 Sep 22. Chembiochem. 2022. PMID: 34416067 Free PMC article. Review.
Cited by
-
Orthogonal alkynyl amino acid reporter for selective labeling of bacterial proteomes during infection.Angew Chem Int Ed Engl. 2010 Aug 9;49(34):5970-4. doi: 10.1002/anie.201002050. Angew Chem Int Ed Engl. 2010. PMID: 20632346 Free PMC article. No abstract available.
-
Residue-Specific Incorporation of Noncanonical Amino Acids in Auxotrophic Hosts: Quo Vadis?.Chem Rev. 2025 May 28;125(10):4840-4932. doi: 10.1021/acs.chemrev.4c00280. Epub 2025 May 16. Chem Rev. 2025. PMID: 40378355 Free PMC article. Review.
-
Small molecule probes for plant cell wall polysaccharide imaging.Front Plant Sci. 2012 May 11;3:89. doi: 10.3389/fpls.2012.00089. eCollection 2012. Front Plant Sci. 2012. PMID: 22639673 Free PMC article.
-
Directed Evolution of the Methanosarcina barkeri Pyrrolysyl tRNA/aminoacyl tRNA Synthetase Pair for Rapid Evaluation of Sense Codon Reassignment Potential.Int J Mol Sci. 2021 Jan 18;22(2):895. doi: 10.3390/ijms22020895. Int J Mol Sci. 2021. PMID: 33477414 Free PMC article.
-
A FRET-based fluorogenic phosphine for live-cell imaging with the Staudinger ligation.Angew Chem Int Ed Engl. 2008;47(13):2394-7. doi: 10.1002/anie.200704847. Angew Chem Int Ed Engl. 2008. PMID: 18306205 Free PMC article. No abstract available.
References
-
- Budisa N. Angew. Chem. Int. Ed. 2004;43:6426–6463. - PubMed
-
- Link A. J., Mock M. L., Tirrell D. A. Curr. Opin. Biotechnol. 2003;14:603–609. - PubMed
-
- Kiick K. L., van Hest J. C. M., Tirrell D. A. Angew. Chem. Int. Ed. 2000;39:2148–2152. - PubMed
-
- Kiick K. L., Weberskirch R., Tirrell D. A. FEBS Lett. 2001;502:25–30. - PubMed
-
- Tang Y., Tirrell D. A. J. Am. Chem. Soc. 2001;123:11089–11090. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

