Just one cross appears capable of dramatically altering the population biology of a eukaryotic pathogen like Toxoplasma gondii
- PMID: 16801557
- PMCID: PMC1502489
- DOI: 10.1073/pnas.0510319103
Just one cross appears capable of dramatically altering the population biology of a eukaryotic pathogen like Toxoplasma gondii
Abstract
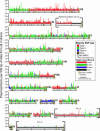
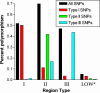
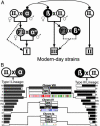
Toxoplasma gondii, an obligate intracellular protozoan of the phylum Apicomplexa, is estimated to infect over a billion people worldwide as well as a great many other mammalian and avian hosts. Despite this ubiquity, the vast majority of human infections in Europe and North America are thought to be due to only three genotypes. Using a genome-wide analysis of single-nucleotide polymorphisms, we have constructed a genealogy for these three lines. The data indicate that types I and III are second- and first-generation offspring, respectively, of a cross between a type II strain and one of two ancestral strains. An extant T. gondii strain (P89) appears to be the modern descendant of the non-type II parent of type III, making the full genealogy of the type III clonotype known. The simplicity of this family tree demonstrates that even a single cross can lead to the emergence and dominance of a new clonal genotype that completely alters the population biology of a sexual pathogen.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

References
-
- Tibayrenc M., Ayala F. J. Trends Parasitol. 2002;18:405–410. - PubMed
-
- Webster I. P. Parasitology. 1994;108:407–411. - PubMed
-
- Aspinall T. V., Guy E. C., Roberts K. E., Joynson D. H., Hyde J. E., Sims P. F. Int. J. Parasitol. 2003;33:97–103. - PubMed
-
- Ajzenberg D., Banuls A. L., Su C., Dumetre A., Demar M., Carme B., Darde M. L. Int. J. Parasitol. 2004;34:1185–1196. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R01 AI036629/AI/NIAID NIH HHS/United States
- AI21423/AI/NIAID NIH HHS/United States
- AI045806/AI/NIAID NIH HHS/United States
- F32 AI060306/AI/NIAID NIH HHS/United States
- R01 AI021423/AI/NIAID NIH HHS/United States
- R37 AI021423/AI/NIAID NIH HHS/United States
- R21 AI045806/AI/NIAID NIH HHS/United States
- AI41014/AI/NIAID NIH HHS/United States
- Z01 AI005093/ImNIH/Intramural NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AI045806/AI/NIAID NIH HHS/United States
- AI05093/AI/NIAID NIH HHS/United States
- F32AI60306/AI/NIAID NIH HHS/United States
- R01 AI041014/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

