Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance
- PMID: 16802858
- PMCID: PMC1484498
- DOI: 10.1371/journal.pbio.0040242
Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance
Abstract
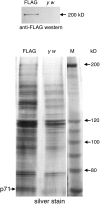
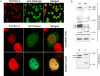
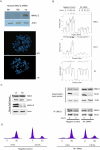
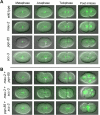
Saccharomyces cerevisiae Scc2 binds Scc4 to form an essential complex that loads cohesin onto chromosomes. The prevalence of Scc2 orthologs in eukaryotes emphasizes a conserved role in regulating sister chromatid cohesion, but homologs of Scc4 have not hitherto been identified outside certain fungi. Some metazoan orthologs of Scc2 were initially identified as developmental gene regulators, such as Drosophila Nipped-B, a regulator of cut and Ultrabithorax, and delangin, a protein mutant in Cornelia de Lange syndrome. We show that delangin and Nipped-B bind previously unstudied human and fly orthologs of Caenorhabditis elegans MAU-2, a non-axis-specific guidance factor for migrating cells and axons. PSI-BLAST shows that Scc4 is evolutionarily related to metazoan MAU-2 sequences, with the greatest homology evident in a short N-terminal domain, and protein-protein interaction studies map the site of interaction between delangin and human MAU-2 to the N-terminal regions of both proteins. Short interfering RNA knockdown of human MAU-2 in HeLa cells resulted in precocious sister chromatid separation and in impaired loading of cohesin onto chromatin, indicating that it is functionally related to Scc4, and RNAi analyses show that MAU-2 regulates chromosome segregation in C. elegans embryos. Using antisense morpholino oligonucleotides to knock down Xenopus tropicalis delangin or MAU-2 in early embryos produced similar patterns of retarded growth and developmental defects. Our data show that sister chromatid cohesion in metazoans involves the formation of a complex similar to the Scc2-Scc4 interaction in the budding yeast. The very high degree of sequence conservation between Scc4 homologs in complex metazoans is consistent with increased selection pressure to conserve additional essential functions, such as regulation of cell and axon migration during development.
Figures



Comment in
-
Ancient protein partners take on additional roles in multicellular animals.PLoS Biol. 2006 Aug;4(8):e266. doi: 10.1371/journal.pbio.0040266. Epub 2006 Jul 4. PLoS Biol. 2006. PMID: 20076621 Free PMC article. No abstract available.
References
-
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. - PubMed
-
- Losada A, Hirano T. Dynamic molecular linkers of the genome: The first decade of SMC proteins. Genes Dev. 2005;19:1269–1287. - PubMed
-
- Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. - PubMed
-
- Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. - PubMed
-
- Ivanov D, Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

