A previously unidentified activity of yeast and mouse RNA:pseudouridine synthases 1 (Pus1p) on tRNAs
- PMID: 16804160
- PMCID: PMC1524882
- DOI: 10.1261/rna.100806
A previously unidentified activity of yeast and mouse RNA:pseudouridine synthases 1 (Pus1p) on tRNAs
Abstract
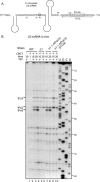
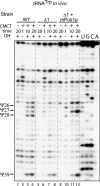
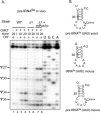
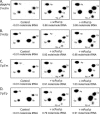
Mouse pseudouridine synthase 1 (mPus1p) was the first vertebrate RNA:pseudouridine synthase that was cloned and characterized biochemically. The mPus1p was previously found to catalyze Psi formation at positions 27, 28, 34, and 36 in in vitro produced yeast and human tRNAs. On the other hand, the homologous Saccharomyces cerevisiae scPus1p protein was shown to modify seven uridine residues in tRNAs (26, 27, 28, 34, 36, 65, and 67) and U44 in U2 snRNA. In this work, we expressed mPus1p in yeast cells lacking scPus1p and studied modification of U2 snRNA and several yeast tRNAs. Our data showed that, in these in vivo conditions, the mouse enzyme efficiently modifies yeast U2 snRNA at position 44 and tRNAs at positions 27, 28, 34, and 36. However, a tRNA:Psi26-synthase activity of mPus1p was not observed. Furthermore, we found that both scPus1p and mPus1p, in vivo and in vitro, have a previously unidentified activity at position 1 in cytoplasmic tRNAArg(ACG). This modification can take place in mature tRNA, as well as in pre-tRNAs with 5' and/or 3' extensions. Thus, we identified the protein carrying one of the last missing yeast tRNA:Psi synthase activities. In addition, our results reveal an additional activity of mPus1p at position 30 in tRNA that scPus1p does not possess.
Figures




References
-
- Adams A., Gottschling D.E., Kaiser C.A., Stearns T. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997.
-
- Ansmant I., Motorin I. Identification of RNA modification enzymes using sequence homology. Mol. Biol. (Mosk.) 2001;35:248–267. - PubMed
-
- Auffinger P., Westhof E. Effects of pseudouridylation on tRNA hydration and dynamics: A theoretical approach. In: Grosjean H., Benne R., editors. The modification and editing of RNA. ASM Press; Washington, DC: 1998. pp. 103–112.
-
- Auxilien S., Crain P.F., Trewyn R.W., Grosjean H. Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA. J. Mol. Biol. 1996;262:437–458. - PubMed
-
- Bakin A., Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center—Analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
