Progression of the cell cycle through mitosis leads to abortion of nascent transcripts
- PMID: 1680567
- PMCID: PMC2755073
- DOI: 10.1016/0092-8674(91)90182-x
Progression of the cell cycle through mitosis leads to abortion of nascent transcripts
Abstract
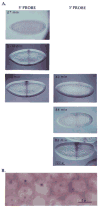
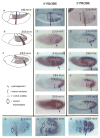
Nascent transcripts of the Drosophila Ubx gene were detected by in situ hybridization. Following onset of expression, the progress of RNA polymerase (1.4 kb/min) across the gene was visualized as the successive appearance of hybridization signals from different positions within the transcription unit. Nascent transcripts disappeared at mitosis. Hybridization signals reappeared in the next cell cycle first with a 5' probe, and later, following a delay consistent with the transcription rate, with a 3' probe. Nascent transcripts were observed continuously in expressing cells of a mutant embryo in which cells are blocked in interphase. We conclude that progression through mitosis causes abortion of nascent transcripts and suggest that periodic abortion of transcription contributes to regulation of expression of large genes.
Figures



References
-
- Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Berlin, Heidelberg, New York, Tokyo: Springer-Verlag; 1985.
-
- Cisek L, Corden J. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature. 1989;339:679–684. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

