Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex
- PMID: 16805913
- PMCID: PMC1524980
- DOI: 10.1186/1476-4598-5-26
Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex
Abstract
Background: Disrupting the balance of histone lysine methylation alters the expression of genes involved in tumorigenesis including proto-oncogenes and cell cycle regulators. Methylation of lysine residues is commonly catalyzed by a family of proteins that contain the SET domain. Here, we report the identification and characterization of the SET domain-containing protein, Smyd2.
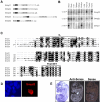
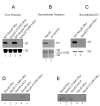
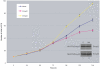
Results: Smyd2 mRNA is most highly expressed in heart and brain tissue, as demonstrated by northern analysis and in situ hybridization. Over-expressed Smyd2 localizes to the cytoplasm and the nucleus in 293T cells. Although accumulating evidence suggests that methylation of histone 3, lysine 36 (H3K36) is associated with actively transcribed genes, we show that the SET domain of Smyd2 mediates H3K36 dimethylation and that Smyd2 represses transcription from an SV40-luciferase reporter. Smyd2 associates specifically with the Sin3A histone deacetylase complex, which was recently linked to H3K36 methylation within the coding regions of active genes in yeast. Finally, we report that exogenous expression of Smyd2 suppresses cell proliferation.
Conclusion: We propose that Sin3A-mediated deacetylation within the coding regions of active genes is directly linked to the histone methyltransferase activity of Smyd2. Moreover, Smyd2 appears to restrain cell proliferation, likely through direct modulation of chromatin structure.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

