Cooperative fluctuations point to the dimerization interface of p53 core domain
- PMID: 16807229
- PMCID: PMC1483080
- DOI: 10.1529/biophysj.106.077800
Cooperative fluctuations point to the dimerization interface of p53 core domain
Abstract
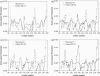
Elastic network models are used for investigation of the p53 core domain functional dynamics. Global modes of motion indicate high positive correlations for residue fluctuations across the A-B interface, which are not observed at the B-C interface. Major hinge formation is observed at the A-B interface upon dimerization indicating stability of the A-B dimer. These findings imply A-B as the native dimerization interface, whereas B-C is the crystal interface. The A-B dimer exhibits an opening-closing motion about DNA, supporting the previously suggested clamp-like model of nonspecific DNA binding followed by diffusion. Monomer A has limited positive correlations with DNA, while monomer B exhibits high positive correlations with DNA in the functionally significant slow modes. Thus, monomer B might seem to maintain the stability of the dimer-DNA complex by forming the relatively fixed arm of the dimer clamp, whereas the other arm of the clamp, monomer A, might allow sliding via continuous association/dissociation mechanisms.
Figures
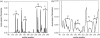
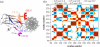


Similar articles
-
NMR spectroscopy reveals the solution dimerization interface of p53 core domains bound to their consensus DNA.J Biol Chem. 2001 Dec 28;276(52):49020-7. doi: 10.1074/jbc.M107516200. Epub 2001 Oct 17. J Biol Chem. 2001. PMID: 11606582
-
Structure of the p53 core domain dimer bound to DNA.J Biol Chem. 2006 Jul 21;281(29):20494-502. doi: 10.1074/jbc.M603634200. Epub 2006 May 22. J Biol Chem. 2006. PMID: 16717092
-
Tandem dimerization of the human p53 tetramerization domain stabilizes a primary dimer intermediate and dramatically enhances its oligomeric stability.J Mol Biol. 2007 Jan 26;365(4):1217-31. doi: 10.1016/j.jmb.2006.10.051. Epub 2006 Oct 21. J Mol Biol. 2007. PMID: 17113101
-
Comparison of the protein-protein interfaces in the p53-DNA crystal structures: towards elucidation of the biological interface.Proc Natl Acad Sci U S A. 2005 Mar 15;102(11):3988-93. doi: 10.1073/pnas.0500215102. Epub 2005 Feb 28. Proc Natl Acad Sci U S A. 2005. PMID: 15738397 Free PMC article.
-
Crystal structure of a p53 core tetramer bound to DNA.Oncogene. 2009 Jan 22;28(3):325-33. doi: 10.1038/onc.2008.400. Epub 2008 Nov 3. Oncogene. 2009. PMID: 18978813 Free PMC article.
Cited by
-
Comparative Study of Elastic Network Model and Protein Contact Network for Protein Complexes: The Hemoglobin Case.Biomed Res Int. 2017;2017:2483264. doi: 10.1155/2017/2483264. Epub 2017 Jan 22. Biomed Res Int. 2017. PMID: 28243596 Free PMC article.
-
Influence of oligomerization on the dynamics of G-protein coupled receptors as assessed by normal mode analysis.Proteins. 2008 May 1;71(2):575-86. doi: 10.1002/prot.21787. Proteins. 2008. PMID: 17963239 Free PMC article.
-
Changes in dynamics upon oligomerization regulate substrate binding and allostery in amino acid kinase family members.PLoS Comput Biol. 2011 Sep;7(9):e1002201. doi: 10.1371/journal.pcbi.1002201. Epub 2011 Sep 29. PLoS Comput Biol. 2011. PMID: 21980279 Free PMC article.
-
Modulation of Toroidal Proteins Dynamics in Favor of Functional Mechanisms upon Ligand Binding.Biophys J. 2020 Apr 7;118(7):1782-1794. doi: 10.1016/j.bpj.2020.01.046. Epub 2020 Feb 18. Biophys J. 2020. PMID: 32130874 Free PMC article.
-
Probing potential binding modes of the p53 tetramer to DNA based on the symmetries encoded in p53 response elements.Nucleic Acids Res. 2007;35(22):7733-47. doi: 10.1093/nar/gkm890. Epub 2007 Nov 5. Nucleic Acids Res. 2007. PMID: 17986463 Free PMC article.
References
-
- el-Diery, W. S. 1998. Regulation of p53 downstream genes. Semin. Cancer Biol. 8:345–357. - PubMed
-
- Ryan, K. M., A. C. Phillips, and K. H. Vousden. 2001. Regulation and function of the p53 tumor suppressor protein. Curr. Opin. Cell Biol. 13:332–337. - PubMed
-
- Hofseth, L. J., S. P. Hussain, and C. C. Harris. 2004. p53: 25 years after its discovery. Trends Pharmacol. Sci. 25:117–181. - PubMed
-
- Donehower, L. A., M. Harvey, B. L. Slagle, M. J. McArthur, C. A. Montgomery, J. S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumors. Nature. 356:215–221. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

