Promiscuous mismatch extension by human DNA polymerase lambda
- PMID: 16807316
- PMCID: PMC1904104
- DOI: 10.1093/nar/gkl377
Promiscuous mismatch extension by human DNA polymerase lambda
Abstract
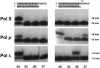
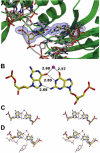
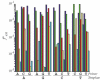
DNA polymerase lambda (Pol lambda) is one of several DNA polymerases suggested to participate in base excision repair (BER), in repair of broken DNA ends and in translesion synthesis. It has been proposed that the nature of the DNA intermediates partly determines which polymerase is used for a particular repair reaction. To test this hypothesis, here we examine the ability of human Pol lambda to extend mismatched primer-termini, either on 'open' template-primer substrates, or on its preferred substrate, a 1 nt gapped-DNA molecule having a 5'-phosphate. Interestingly, Pol lambda extended mismatches with an average efficiency of approximately 10(-2) relative to matched base pairs. The match and mismatch extension catalytic efficiencies obtained on gapped molecules were approximately 260-fold higher than on template-primer molecules. A crystal structure of Pol lambda in complex with a single-nucleotide gap containing a dG.dGMP mismatch at the primer-terminus (2.40 A) suggests that, at least for certain mispairs, Pol lambda is unable to differentiate between matched and mismatched termini during the DNA binding step, thus accounting for the relatively high efficiency of mismatch extension. This property of Pol lambda suggests a potential role as a 'mismatch extender' during non-homologous end joining (NHEJ), and possibly during translesion synthesis.
Figures
Similar articles
-
DNA polymerase X of African swine fever virus: insertion fidelity on gapped DNA substrates and AP lyase activity support a role in base excision repair of viral DNA.J Mol Biol. 2003 Mar 7;326(5):1403-12. doi: 10.1016/s0022-2836(03)00019-6. J Mol Biol. 2003. PMID: 12595253
-
Efficiency and fidelity of human DNA polymerases λ and β during gap-filling DNA synthesis.DNA Repair (Amst). 2011 Jan 2;10(1):24-33. doi: 10.1016/j.dnarep.2010.09.005. Epub 2010 Oct 20. DNA Repair (Amst). 2011. PMID: 20961817 Free PMC article.
-
Influence of DNA structure on DNA polymerase beta active site function: extension of mutagenic DNA intermediates.J Biol Chem. 2004 Jul 23;279(30):31921-9. doi: 10.1074/jbc.M404016200. Epub 2004 May 15. J Biol Chem. 2004. PMID: 15145936
-
Ubiquitylation of DNA polymerase λ.FEBS Lett. 2011 Sep 16;585(18):2826-30. doi: 10.1016/j.febslet.2011.03.069. Epub 2011 Apr 8. FEBS Lett. 2011. PMID: 21486570 Review.
-
20 years of DNA Polymerase μ, the polymerase that still surprises.FEBS J. 2021 Dec;288(24):7230-7242. doi: 10.1111/febs.15852. Epub 2021 Apr 25. FEBS J. 2021. PMID: 33786971 Review.
Cited by
-
Involvement of AtPolλ in the repair of high salt- and DNA cross-linking agent-induced double strand breaks in Arabidopsis.Plant Physiol. 2013 Jun;162(2):1195-210. doi: 10.1104/pp.113.219022. Epub 2013 May 9. Plant Physiol. 2013. PMID: 23660835 Free PMC article.
-
The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins.Nucleic Acids Res. 2006;34(17):4731-42. doi: 10.1093/nar/gkl465. Epub 2006 Sep 13. Nucleic Acids Res. 2006. PMID: 16971464 Free PMC article.
-
A role for DNA polymerase mu in the emerging DJH rearrangements of the postgastrulation mouse embryo.Mol Cell Biol. 2009 Mar;29(5):1266-75. doi: 10.1128/MCB.01518-08. Epub 2008 Dec 22. Mol Cell Biol. 2009. PMID: 19103746 Free PMC article.
-
Pif1 family helicases promote mutation avoidance during DNA replication.Nucleic Acids Res. 2022 Dec 9;50(22):12844-12855. doi: 10.1093/nar/gkac1127. Nucleic Acids Res. 2022. PMID: 36533450 Free PMC article.
-
How asymmetric DNA replication achieves symmetrical fidelity.Nat Struct Mol Biol. 2021 Dec;28(12):1020-1028. doi: 10.1038/s41594-021-00691-6. Epub 2021 Dec 9. Nat Struct Mol Biol. 2021. PMID: 34887558 Free PMC article.
References
-
- Bebenek K., Kunkel T.A. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. - PubMed
-
- Burgers P.M., Koonin E.V., Bruford E., Blanco L., Burtis K.C., Christman M.F., Copeland W.C., Friedberg E.C., Hanaoka F., Hinkle D.C., et al. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 2001;276:43487–43490. - PubMed
-
- Oliveros M., Yanez R.J., Salas M.L., Salas J., Vinuela E., Blanco L. Characterization of an African swine fever virus 20 kDa DNA polymerase envolved in DNA repair. J. Biol. Chem. 1997;272:30899–30910. - PubMed
-
- Garcia-Diaz M., Dominguez O., Lopez-Fernandez L.A., de Lera L.T., Saniger M.L., Ruiz J.F., Parraga M., Garcia-Ortiz M.J., Kirchhoff T., del Mazo J., et al. DNA polymerase lambda (Pol lambda), a novel eukaryotic DNA polymerase with a potencial role in meiosis. J. Mol. Biol. 2000;301:851–867. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

