Consistent effects of TSG101 genetic variability on multiple outcomes of exposure to human immunodeficiency virus type 1
- PMID: 16809281
- PMCID: PMC1489017
- DOI: 10.1128/jvi.00094-06
Consistent effects of TSG101 genetic variability on multiple outcomes of exposure to human immunodeficiency virus type 1
Abstract
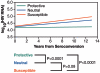
Tumor susceptibility gene 101 (TSG101) encodes a host cellular protein that is appropriated by human immunodeficiency virus type 1 (HIV-1) in the budding process of viral particles from infected cells. Variation in the coding or noncoding regions of the gene could potentially affect the degree of TSG101-mediated release of viral particles. While the coding regions of the gene were found to lack nonsynonymous variants, two polymorphic sites in the TSG101 5' area were identified that were associated with the rate of AIDS progression among Caucasians. These single-nucleotide polymorphisms (SNPs), located at positions -183 and +181 relative to the translation start, specify three haplotypes termed A, B, and C, which occur at frequencies of 67%, 21%, and 12%, respectively. Haplotype C is associated with relatively rapid AIDS progression, while haplotype B is associated with slower disease progression. Both effects were dominant over the intermediate haplotype A. The haplotypes also demonstrated parallel effects on the rate of CD4 T-cell depletion and viral load increase over time, as well as a possible influence on HIV-1 infection. The data raise the hypothesis that noncoding variation in TSG101 affects the efficiency of TSG101-mediated release of viral particles from infected cells, thereby altering levels of plasma viral load and subsequent disease progression.
Figures



References
-
- Babst, M., G. Odorizzi, E. J. Estepa, and S. D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1:248-258. - PubMed
-
- Bleiber, G., M. May, R. Martinez, P. Meylan, J. Ott, J. S. Beckmann, and A. Telenti. 2005. Use of a combined ex vivo/in vivo population approach for screening of human genes involved in the human immunodeficiency virus type 1 life cycle for variants influencing disease progression. J. Virol. 79:12674-12680. - PMC - PubMed
-
- Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. - PMC - PubMed
-
- Burgdorf, S., P. Leister, and K. H. Scheidtmann. 2004. TSG101 interacts with apoptosis-antagonizing transcription factor and enhances androgen receptor-mediated transcription by promoting its monoubiquitination. J. Biol. Chem. 279:17524-17534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

