Role of invariant Thr80 in human immunodeficiency virus type 1 protease structure, function, and viral infectivity
- PMID: 16809296
- PMCID: PMC1489026
- DOI: 10.1128/JVI.01900-05
Role of invariant Thr80 in human immunodeficiency virus type 1 protease structure, function, and viral infectivity
Abstract
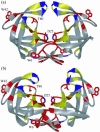
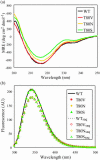
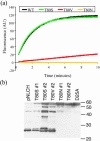
Sequence variability associated with human immunodeficiency virus type 1 (HIV-1) is useful for inferring structural and/or functional constraints at specific residues within the viral protease. Positions that are invariant even in the presence of drug selection define critically important residues for protease function. While the importance of conserved active-site residues is easily understood, the role of other invariant residues is not. This work focuses on invariant Thr80 at the apex of the P1 loop of HIV-1, HIV-2, and simian immunodeficiency virus protease. In a previous study, we postulated, on the basis of a molecular dynamics simulation of the unliganded protease, that Thr80 may play a role in the mobility of the flaps of protease. In the present study, both experimental and computational methods were used to study the role of Thr80 in HIV protease. Three protease variants (T80V, T80N, and T80S) were examined for changes in structure, dynamics, enzymatic activity, affinity for protease inhibitors, and viral infectivity. While all three variants were structurally similar to the wild type, only T80S was functionally similar. Both T80V and T80N had decreased the affinity for saquinavir. T80V significantly decreased the ability of the enzyme to cleave a peptide substrate but maintained infectivity, while T80N abolished both activity and viral infectivity. Additionally, T80N decreased the conformational flexibility of the flap region, as observed by simulations of molecular dynamics. Taken together, these data indicate that HIV-1 protease functions best when residue 80 is a small polar residue and that mutations to other amino acids significantly impair enzyme function, possibly by affecting the flexibility of the flap domain.
Figures


Similar articles
-
Structural basis for distinctions between substrate and inhibitor specificities for feline immunodeficiency virus and human immunodeficiency virus proteases.J Virol. 2003 Jun;77(12):6589-600. doi: 10.1128/jvi.77.12.6589-6600.2003. J Virol. 2003. PMID: 12767979 Free PMC article.
-
Domain flexibility in retroviral proteases: structural implications for drug resistant mutations.Biochemistry. 1998 Feb 24;37(8):2607-21. doi: 10.1021/bi9716074. Biochemistry. 1998. PMID: 9485411
-
Structural role of the 30's loop in determining the ligand specificity of the human immunodeficiency virus protease.Biochemistry. 1998 Aug 4;37(31):10928-36. doi: 10.1021/bi980784h. Biochemistry. 1998. PMID: 9692985
-
Qualitative study of drug resistance in retroviral protease using structural modeling and site-directed mutagenesis.Methods Enzymol. 1994;241:385-94. doi: 10.1016/0076-6879(94)41075-5. Methods Enzymol. 1994. PMID: 7854190 Review. No abstract available.
-
Probing structure-function relationships in human immunodeficiency virus type 1 protease via molecular dynamics simulation.Methods Enzymol. 1994;241:178-95. doi: 10.1016/0076-6879(94)41065-8. Methods Enzymol. 1994. PMID: 7854178 Review.
Cited by
-
Differential Flap Dynamics in Wild-type and a Drug Resistant Variant of HIV-1 Protease Revealed by Molecular Dynamics and NMR Relaxation.J Chem Theory Comput. 2012 Oct 9;8(10):3452-3462. doi: 10.1021/ct300076y. Epub 2012 Apr 17. J Chem Theory Comput. 2012. PMID: 23144597 Free PMC article.
-
Structural Adaptation of Darunavir Analogues against Primary Mutations in HIV-1 Protease.ACS Infect Dis. 2019 Feb 8;5(2):316-325. doi: 10.1021/acsinfecdis.8b00336. Epub 2018 Dec 31. ACS Infect Dis. 2019. PMID: 30543749 Free PMC article.
-
Caught in the Act: the 1.5 A resolution crystal structures of the HIV-1 protease and the I54V mutant reveal a tetrahedral reaction intermediate.Biochemistry. 2007 Dec 25;46(51):14854-64. doi: 10.1021/bi700822g. Epub 2007 Dec 4. Biochemistry. 2007. PMID: 18052235 Free PMC article.
-
A synergy of activity, stability, and inhibitor-interaction of HIV-1 protease mutants evolved under drug-pressure.Protein Sci. 2021 Mar;30(3):571-582. doi: 10.1002/pro.4013. Epub 2020 Dec 22. Protein Sci. 2021. PMID: 33314454 Free PMC article.
-
Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.J Mol Biol. 2008 Aug 1;381(1):102-15. doi: 10.1016/j.jmb.2008.05.062. Epub 2008 Jul 1. J Mol Biol. 2008. PMID: 18597780 Free PMC article.
References
-
- Ala, P. J., E. E. Huston, R. M. Klabe, P. K. Jadhav, P. Y. Lam, and C. H. Chang. 1998. Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities. Biochemistry 37:15042-15049. - PubMed
-
- Ariyoshi, K., M. Matsuda, H. Miura, S. Tateishi, K. Yamada, and W. Sugiura. 2003. Patterns of point mutations associated with antiretroviral drug treatment failure in CRF01_AE (subtype E) infection differ from subtype B infection. J. Acquir. Immune Defic. Syndr. 33:336-342. - PubMed
-
- Baldwin, E. T., T. N. Bhat, B. Liu, N. Pattabiraman, and J. W. Erickson. 1995. Structural basis of drug resistance for the V82A mutant of HIV-1 proteinase. Nat. Struct. Biol. 2:244-249. - PubMed
-
- Beasley, J. R., D. F. Doyle, L. Chen, C. D. S., B. R. Fine, and G. J. Peilak. 2002. Searching for quantitative entropy-enthalpy compensation among protein variants. Proteins Struct. Funct. Genet. 49:398-402. - PubMed
-
- Blasie, C. A., and J. M. Berg. 2004. Entropy-enthalpy compensation in ionic interactions probed in a zinc finger peptide. Biochemistry 43:10600-10604. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

