Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol
- PMID: 16809311
- PMCID: PMC1489054
- DOI: 10.1128/JVI.02195-05
Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol
Abstract
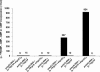
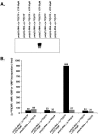
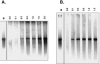
Noroviruses (Caliciviridae) are RNA viruses with a single-stranded, positive-oriented polyadenylated genome. To date, little is known about the replication strategy of norovirus, a so-far noncultivable virus. We have examined the initiation of replication of the norovirus genome in vitro, using the active norovirus RNA-dependent RNA polymerase (3D(pol)), homopolymeric templates, and synthetic subgenomic or antisubgenomic RNA. Initiation of RNA synthesis on homopolymeric templates as well as replication of subgenomic polyadenylated RNA was strictly primer dependent. In this context and as observed for other enteric RNA viruses, i.e., poliovirus, a protein-primed initiation of RNA synthesis after elongation of the VPg by norovirus 3D(pol) was postulated. To address this question, norovirus VPg was expressed in Escherichia coli and purified. Incubation of VPg with norovirus 3D(pol) generated VPg-poly(U), which primed the replication of subgenomic polyadenylated RNA. In contrast, replication of antisubgenomic RNA was not primer dependent, nor did it depend on a leader sequence, as evidenced by deletion analysis of the 3' termini of subgenomic and antisubgenomic RNA. On nonpolyadenylated RNA, i.e., antisubgenomic RNA, norovirus 3D(pol) initiated RNA synthesis de novo and terminated RNA synthesis by a poly(C) stretch. Interestingly, on poly(C) RNA templates, norovirus 3D(pol) initiated RNA synthesis de novo in the presence of high concentrations of GTP. We propose a novel model for initiation of replication of the norovirus genome by 3D(pol), with a VPg-protein-primed initiation of replication of polyadenylated genomic RNA and a de novo initiation of replication of antigenomic RNA.
Figures








Similar articles
-
Characterization of norovirus 3Dpol RNA-dependent RNA polymerase activity and initiation of RNA synthesis.J Gen Virol. 2006 Sep;87(Pt 9):2621-2630. doi: 10.1099/vir.0.81802-0. J Gen Virol. 2006. PMID: 16894201
-
Murine norovirus-1 3Dpol exhibits RNA-dependent RNA polymerase activity and nucleotidylylates on Tyr of the VPg.J Gen Virol. 2010 Jul;91(Pt 7):1713-22. doi: 10.1099/vir.0.020461-0. Epub 2010 Mar 10. J Gen Virol. 2010. PMID: 20219896
-
Flock house virus RNA polymerase initiates RNA synthesis de novo and possesses a terminal nucleotidyl transferase activity.PLoS One. 2014 Jan 23;9(1):e86876. doi: 10.1371/journal.pone.0086876. eCollection 2014. PLoS One. 2014. PMID: 24466277 Free PMC article.
-
Protein Nucleotidylylation in +ssRNA Viruses.Viruses. 2021 Aug 5;13(8):1549. doi: 10.3390/v13081549. Viruses. 2021. PMID: 34452414 Free PMC article. Review.
-
Cis-active RNA elements (CREs) and picornavirus RNA replication.Virus Res. 2009 Feb;139(2):240-52. doi: 10.1016/j.virusres.2008.07.027. Epub 2008 Sep 20. Virus Res. 2009. PMID: 18773930 Free PMC article. Review.
Cited by
-
The importance of inter- and intramolecular base pairing for translation reinitiation on a eukaryotic bicistronic mRNA.Genes Dev. 2009 Feb 1;23(3):331-44. doi: 10.1101/gad.507609. Genes Dev. 2009. PMID: 19204118 Free PMC article.
-
Two alternative ways of start site selection in human norovirus reinitiation of translation.J Biol Chem. 2014 Apr 25;289(17):11739-11754. doi: 10.1074/jbc.M114.554030. Epub 2014 Mar 5. J Biol Chem. 2014. PMID: 24599949 Free PMC article.
-
The Disorderly Nature of Caliciviruses.Viruses. 2024 Aug 19;16(8):1324. doi: 10.3390/v16081324. Viruses. 2024. PMID: 39205298 Free PMC article. Review.
-
Polyprotein processing and intermolecular interactions within the viral replication complex spatially and temporally control norovirus protease activity.J Biol Chem. 2019 Mar 15;294(11):4259-4271. doi: 10.1074/jbc.RA118.006780. Epub 2019 Jan 15. J Biol Chem. 2019. PMID: 30647130 Free PMC article.
-
Structure and Function of Caliciviral RNA Polymerases.Viruses. 2017 Nov 6;9(11):329. doi: 10.3390/v9110329. Viruses. 2017. PMID: 29113097 Free PMC article. Review.
References
-
- Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. - PubMed
-
- Green, K. Y., R. M. Chanock, and A. Z. Kapikian. 2001. Human caliciviruses. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

