Hypersusceptibility to substrate analogs conferred by mutations in human immunodeficiency virus type 1 reverse transcriptase
- PMID: 16809322
- PMCID: PMC1489025
- DOI: 10.1128/JVI.00322-06
Hypersusceptibility to substrate analogs conferred by mutations in human immunodeficiency virus type 1 reverse transcriptase
Abstract
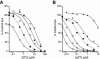
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) contains four structural motifs (A, B, C, and D) that are conserved in polymerases from diverse organisms. Motif B interacts with the incoming nucleotide, the template strand, and key active-site residues from other motifs, suggesting that motif B is an important determinant of substrate specificity. To examine the functional role of this region, we performed "random scanning mutagenesis" of 11 motif B residues and screened replication-competent mutants for altered substrate analog sensitivity in culture. Single amino acid replacements throughout the targeted region conferred resistance to lamivudine and/or hypersusceptibility to zidovudine (AZT). Substitutions at residue Q151 increased the sensitivity of HIV-1 to multiple nucleoside analogs, and a subset of these Q151 variants was also hypersusceptible to the pyrophosphate analog phosphonoformic acid (PFA). Other AZT-hypersusceptible mutants were resistant to PFA and are therefore phenotypically similar to PFA-resistant variants selected in vitro and in infected patients. Collectively, these data show that specific amino acid replacements in motif B confer broad-spectrum hypersusceptibility to substrate analog inhibitors. Our results suggest that motif B influences RT-deoxynucleoside triphosphate interactions at multiple steps in the catalytic cycle of polymerization.
Figures

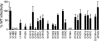
Similar articles
-
Relationship between 3'-azido-3'-deoxythymidine resistance and primer unblocking activity in foscarnet-resistant mutants of human immunodeficiency virus type 1 reverse transcriptase.J Virol. 2003 Jun;77(11):6127-37. doi: 10.1128/jvi.77.11.6127-6137.2003. J Virol. 2003. PMID: 12743270 Free PMC article.
-
Phosphorylation of AZT-resistant human immunodeficiency virus type 1 reverse transcriptase by casein kinase II in vitro: effects on inhibitor sensitivity.Biochem Biophys Res Commun. 2000 Aug 18;275(1):26-32. doi: 10.1006/bbrc.2000.3251. Biochem Biophys Res Commun. 2000. PMID: 10944435
-
Long-term foscarnet therapy remodels thymidine analogue mutations and alters resistance to zidovudine and lamivudine in HIV-1.Antivir Ther. 2007;12(3):335-43. Antivir Ther. 2007. PMID: 17591023
-
Structural and biochemical effects of human immunodeficiency virus mutants resistant to non-nucleoside reverse transcriptase inhibitors.Int J Biochem Cell Biol. 2004 Sep;36(9):1735-51. doi: 10.1016/j.biocel.2004.02.026. Int J Biochem Cell Biol. 2004. PMID: 15183341 Review.
-
The lysine 65 residue in HIV-1 reverse transcriptase function and in nucleoside analog drug resistance.Viruses. 2014 Oct 23;6(10):4080-94. doi: 10.3390/v6104080. Viruses. 2014. PMID: 25341667 Free PMC article. Review.
Cited by
-
Extensive mutagenesis of the conserved box E motif in duck hepatitis B virus P protein reveals multiple functions in replication and a common structure with the primer grip in HIV-1 reverse transcriptase.J Virol. 2012 Jun;86(12):6394-407. doi: 10.1128/JVI.00011-12. Epub 2012 Apr 18. J Virol. 2012. PMID: 22514339 Free PMC article.
-
Human immunodeficiency virus types 1 and 2 exhibit comparable sensitivities to Zidovudine and other nucleoside analog inhibitors in vitro.Antimicrob Agents Chemother. 2008 Jan;52(1):329-32. doi: 10.1128/AAC.01004-07. Epub 2007 Oct 29. Antimicrob Agents Chemother. 2008. PMID: 17967913 Free PMC article.
-
Antiretroviral drug resistance in HIV-2: three amino acid changes are sufficient for classwide nucleoside analogue resistance.J Infect Dis. 2009 May 1;199(9):1323-6. doi: 10.1086/597802. J Infect Dis. 2009. PMID: 19358668 Free PMC article.
-
Bioinformatic Annotation of Transposon DNA Processing Genes on the Long-Read Genome Assembly of Caenorhabditis elegans.Bioinform Biol Insights. 2024 Dec 20;18:11779322241304668. doi: 10.1177/11779322241304668. eCollection 2024. Bioinform Biol Insights. 2024. PMID: 39713040 Free PMC article.
-
Conformational States of HIV-1 Reverse Transcriptase for Nucleotide Incorporation vs Pyrophosphorolysis-Binding of Foscarnet.ACS Chem Biol. 2016 Aug 19;11(8):2158-64. doi: 10.1021/acschembio.6b00187. Epub 2016 Jun 6. ACS Chem Biol. 2016. PMID: 27192549 Free PMC article.
References
-
- Arion, D., N. Sluis-Cremer, and M. A. Parniak. 2000. Mechanism by which phosphonoformic acid resistance mutations restore 3′-azido-3′-deoxythymidine (AZT) sensitivity to AZT-resistant HIV-1 reverse transcriptase. J. Biol. Chem. 275:9251-9255. - PubMed
-
- Astatke, M., N. D. Grindley, and C. M. Joyce. 1998. How E. coli DNA polymerase I (Klenow fragment) distinguishes between deoxy- and dideoxynucleotides. J. Mol. Biol. 278:147-165. - PubMed
-
- Charneau, P., G. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241:651-662. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

