Differential binding of replication proteins across the human c-myc replicator
- PMID: 16809765
- PMCID: PMC1592723
- DOI: 10.1128/MCB.02137-05
Differential binding of replication proteins across the human c-myc replicator
Abstract
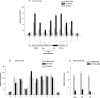
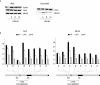
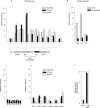
The binding of the prereplication complex proteins Orc1, Orc2, Mcm3, Mcm7, and Cdc6 and the novel DNA unwinding element (DUE) binding protein DUE-B to the endogenous human c-myc replicator was studied by chromatin immunoprecipitation. In G(1)-arrested HeLa cells, Mcm3, Mcm7, and DUE-B were prominent near the DUE, while Orc1 and Orc2 were least abundant near the DUE and more abundant at flanking sites. Cdc6 binding mirrored that of Orc2 in G(1)-arrested cells but decreased in asynchronous or M-phase cells. Similarly, the signals from Orc1, Mcm3, and Mcm7 were at background levels in cells arrested in M phase, whereas Orc2 retained the distribution seen in G(1)-phase cells. Previously shown to cause histone hyperacetylation and delocalization of replication initiation, trichostatin A treatment of cells led to a parallel qualitative change in the distribution of Mcm3, but not Orc2, across the c-myc replicator. Orc2, Mcm3, and DUE-B were also bound at an ectopic c-myc replicator, where deletion of sequences essential for origin activity was associated with the loss of DUE-B binding or the alteration of chromatin structure and loss of Mcm3 binding. These results show that proteins implicated in replication initiation are selectively and differentially bound across the c-myc replicator, dependent on discrete structural elements in DNA or chromatin.
Figures


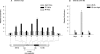


References
-
- Aggarwal, B. D., and B. R. Calvi. 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430:372-376. - PubMed
-
- Aladjem, M. I., L. W. Rodewald, J. L. Kolman, and G. M. Wahl. 1998. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281:1005-1009. - PubMed
-
- Alexandrow, M. G., M. Ritzi, A. Pemov, and J. L. Hamlin. 2002. A potential role for mini-chromosome maintenance (MCM) proteins in initiation at the dihydrofolate reductase replication origin. J. Biol. Chem. 277:2702-2708. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
