Wnt activation and alternative promoter repression of LEF1 in colon cancer
- PMID: 16809766
- PMCID: PMC1592719
- DOI: 10.1128/MCB.00105-06
Wnt activation and alternative promoter repression of LEF1 in colon cancer
Abstract
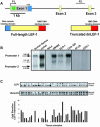
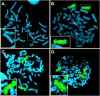
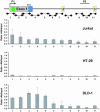
Alternative promoters within the LEF1 locus produce polypeptides of opposing biological activities. Promoter 1 produces full-length LEF-1 protein, which recruits beta-catenin to Wnt target genes. Promoter 2 produces a truncated form that cannot interact with beta-catenin and instead suppresses Wnt regulation of target genes. Here we show that promoter 1 is aberrantly activated in colon cancers because it is a direct target of the Wnt pathway. T-cell factor (TCF)-beta-catenin complexes bind to Wnt response elements in exon 1 and dynamically regulate chromatin acetylation and promoter 1 activity. Promoter 2 is delimited to the intron 2/exon 3 boundary and, like promoter 1, is also directly regulated by TCF-beta-catenin complexes. Promoter 2 is nevertheless silent in colon cancer because an upstream repressor selectively targets the basal promoter leading to destabilized TCF-beta-catenin binding. We conclude that the biological outcome of aberrant LEF1 activation in colon cancer is directed by differential promoter activation and repression.
Figures

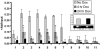



References
-
- Aoki, M., V. Sobek, D. J. Maslyar, A. Hecht, and P. K. Vogt. 2002. Oncogenic transformation by beta-catenin: deletion analysis and characterization of selected target genes. Oncogene 21:6983-6991. - PubMed
-
- Atcha, F. A., J. E. Munguia, T. W. Li, K. Hovanes, and M. L. Waterman. 2003. A new beta-catenin-dependent activation domain in T cell factor. J. Biol. Chem. 278:16169-16175. - PubMed
-
- Batlle, E., J. T. Henderson, H. Beghtel, M. M. van den Born, E. Sancho, G. Huls, J. Meeldijk, J. Robertson, M. van de Wetering, T. Pawson, and H. Clevers. 2002. β-Catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/EphrinB. Cell 111:251-263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
