BAL1 and BBAP are regulated by a gamma interferon-responsive bidirectional promoter and are overexpressed in diffuse large B-cell lymphomas with a prominent inflammatory infiltrate
- PMID: 16809771
- PMCID: PMC1592708
- DOI: 10.1128/MCB.02351-05
BAL1 and BBAP are regulated by a gamma interferon-responsive bidirectional promoter and are overexpressed in diffuse large B-cell lymphomas with a prominent inflammatory infiltrate
Abstract
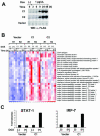
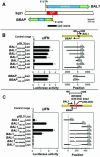
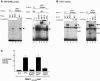
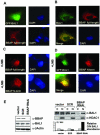
BAL1 is a transcription modulator that is overexpressed in chemoresistant, diffuse large B-cell lymphomas (DLBCLs). BAL1 complexes with a recently described DELTEX family member termed BBAP. Herein, we characterized BAL1 and BBAP expression in primary DLBCL subtypes defined by their comprehensive transcriptional profiles. BAL1 and BBAP were most abundant in lymphomas with a brisk host inflammatory response, designated host response (HR) tumors. Although these DLBCLs include significant numbers of tumor-infiltrating lymphocytes and interdigitating dendritic cells, BAL1 and BBAP were expressed primarily by malignant B cells, prompting speculation that the genes might be induced by host-derived inflammatory mediators such as gamma interferon (IFN-gamma). In fact, IFN-gamma induced BAL1 and BBAP expression in DLBCL cell lines; doxycycline-induced BAL1 also increased the expression of multiple IFN-stimulated genes, directly implicating BAL1 in an IFN signaling pathway. We show that BAL1 and BBAP are located on chromosome 3q21 in a head-to-head orientation and are regulated by a IFN-gamma-responsive bidirectional promoter. BBAP regulates the subcellular localization of BAL1 by a dynamic shuttling mechanism, highlighting the functional requirement for coordinated BBAP and BAL1 expression. IFN-gamma-induced BAL1/BBAP expression contributes to the molecular signature of HR DLBCLs and highlights the interplay between the inflammatory infiltrate and malignant B cells in these tumors.
Figures


Similar articles
-
BAL1/ARTD9 represses the anti-proliferative and pro-apoptotic IFNγ-STAT1-IRF1-p53 axis in diffuse large B-cell lymphoma.J Cell Sci. 2013 May 1;126(Pt 9):1969-80. doi: 10.1242/jcs.118174. Epub 2013 Mar 13. J Cell Sci. 2013. PMID: 23487038
-
BAL1 and its partner E3 ligase, BBAP, link Poly(ADP-ribose) activation, ubiquitylation, and double-strand DNA repair independent of ATM, MDC1, and RNF8.Mol Cell Biol. 2013 Feb;33(4):845-57. doi: 10.1128/MCB.00990-12. Epub 2012 Dec 10. Mol Cell Biol. 2013. PMID: 23230272 Free PMC article.
-
Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response.Blood. 2005 Mar 1;105(5):1851-61. doi: 10.1182/blood-2004-07-2947. Epub 2004 Nov 18. Blood. 2005. PMID: 15550490
-
BCL-6 in diffuse large-cell lymphomas.Important Adv Oncol. 1996:139-48. Important Adv Oncol. 1996. PMID: 8791133 Review. No abstract available.
-
Mediastinal B-cell lymphoma, a lymphoma type with several characteristics unique among diffuse large B-cell lymphomas.Ann Hematol. 2001;80 Suppl 3:B49-53. doi: 10.1007/pl00022789. Ann Hematol. 2001. PMID: 11757707 Review.
Cited by
-
A machine learning heuristic to identify biologically relevant and minimal biomarker panels from omics data.BMC Genomics. 2015;16 Suppl 1(Suppl 1):S2. doi: 10.1186/1471-2164-16-S1-S2. Epub 2015 Jan 15. BMC Genomics. 2015. PMID: 25923811 Free PMC article.
-
Identification of tumor microenvironment-related genes in lower-grade gliomas by mining TCGA database.Transl Cancer Res. 2020 Aug;9(8):4583-4595. doi: 10.21037/tcr-20-1079. Transl Cancer Res. 2020. PMID: 35117823 Free PMC article.
-
HDAC1,2 inhibition impairs EZH2- and BBAP-mediated DNA repair to overcome chemoresistance in EZH2 gain-of-function mutant diffuse large B-cell lymphoma.Oncotarget. 2015 Mar 10;6(7):4863-87. doi: 10.18632/oncotarget.3120. Oncotarget. 2015. PMID: 25605023 Free PMC article.
-
BCL6 modulates tonic BCR signaling in diffuse large B-cell lymphomas by repressing the SYK phosphatase, PTPROt.Blood. 2009 Dec 17;114(26):5315-21. doi: 10.1182/blood-2009-02-204362. Epub 2009 Oct 23. Blood. 2009. PMID: 19855081 Free PMC article.
-
PARP14 is regulated by the PARP9/DTX3L complex and promotes interferon γ-induced ADP-ribosylation.EMBO J. 2024 Jul;43(14):2908-2928. doi: 10.1038/s44318-024-00125-1. Epub 2024 Jun 4. EMBO J. 2024. PMID: 38834852 Free PMC article.
References
-
- Abramson, J. S., and M. A. Shipp. 2005. Advances in the biology and therapy of diffuse large B-cell lymphoma: moving toward a molecularly targeted approach. Blood 106:1164-1174. - PubMed
-
- Aguiar, R. C., K. Takeyama, C. He, K. Kreinbrink, and M. A. Shipp. 2005. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 280:33756-33765. - PubMed
-
- Aguiar, R. C., Y. Yakushijin, S. Kharbanda, R. Salgia, J. A. Fletcher, and M. A. Shipp. 2000. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood 96:4328-4334. - PubMed
-
- Albig, W., P. Kioschis, A. Poustka, K. Meergans, and D. Doenecke. 1997. Human histone gene organization: nonregular arrangement within a large cluster. Genomics 40:314-322. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
