Cell cycle-dependent regulation of Saccharomyces cerevisiae donor preference during mating-type switching by SBF (Swi4/Swi6) and Fkh1
- PMID: 16809780
- PMCID: PMC1592702
- DOI: 10.1128/MCB.02443-05
Cell cycle-dependent regulation of Saccharomyces cerevisiae donor preference during mating-type switching by SBF (Swi4/Swi6) and Fkh1
Abstract
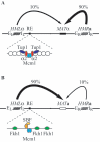
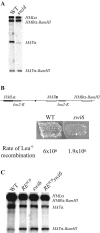
Saccharomyces mating-type switching occurs through a double-strand break-initiated gene conversion event at MAT, using one of two donors located distantly on the same chromosome, HMLalpha and HMRa. MATa cells preferentially choose HMLalpha, a decision that depends on the recombination enhancer (RE) that controls recombination along the left arm of chromosome III. We previously showed that an fhk1Delta mutation reduces HMLalpha usage in MATa cells, but not to the level seen when RE is deleted. We now report that donor preference also depends on binding of the Swi4/Swi6 (SBF) transcription factors to an evolutionarily conserved SCB site within RE. As at other SCB-containing promoters, SBF binds to RE in the G(1) phase. Surprisingly, Fkh1 binds to RE only in G(2), which contrasts with its cell cycle-independent binding to its other target promoters. SBF and Fkh1 define two independent RE activation pathways, as deletion of both Fkh1 and SCB results in nearly complete loss of HML usage in MATa cells. These transcription factors create an epigenetic modification of RE in a fashion that apparently does not involve transcription. In addition, the putative helicase Chl1, previously involved in donor preference, functions in the SBF pathway.
Figures


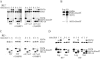
References
-
- Bishop, A. C., J. A. Ubersax, D. T. Petsch, D. P. Matheos, N. S. Gray, J. Blethrow, E. Shimizu, J. Z. Tsien, P. G. Schultz, M. D. Rose, J. L. Wood, D. O. Morgan, and K. M. Shokat. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407:395-401. - PubMed
-
- Breeden, L. 1996. Start-specific transcription in yeast. Curr. Top. Microbiol. Immunol. 208:95-127. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
