Multitasking C2H2 zinc fingers link Zac DNA binding to coordinated regulation of p300-histone acetyltransferase activity
- PMID: 16809786
- PMCID: PMC1592709
- DOI: 10.1128/MCB.02270-05
Multitasking C2H2 zinc fingers link Zac DNA binding to coordinated regulation of p300-histone acetyltransferase activity
Abstract
Zac is a C(2)H(2) zinc finger protein that regulates apoptosis and cell cycle arrest through DNA binding and transactivation. The coactivator proteins p300/CBP enhance transactivation through their histone acetyltransferase (HAT) activity by modulating chromatin structure. Here, we show that p300 increases Zac transactivation in a strictly HAT-dependent manner. Whereas the classic recruitment model proposes that coactivation simply depends on the capacity of the activator to recruit the coactivator, we demonstrate that coordinated binding of Zac zinc fingers and C terminus to p300 regulates HAT function by increasing histone and acetyl coenzyme A affinities and catalytic activity. This concerted regulation of HAT function is mediated via the KIX and CH3 domains of p300 in an interdependent manner. Interestingly, Zac zinc fingers 6 and 7 simultaneously play key roles in DNA binding and p300 regulation. Our findings demonstrate, for the first time, that C(2)H(2) zinc fingers can link DNA binding to HAT signaling and suggest a dynamic role for DNA-binding proteins in the enzymatic control of transcription.
Figures
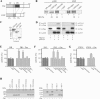
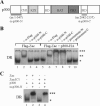
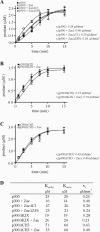
 ) are supershifted by anti-Flag antibodies (
) are supershifted by anti-Flag antibodies (
 ). Anti-p300 antibodies (α-p300-N and α-p300-C) were ineffective in the absence of ectopic p300 (lanes 3 and 4) but strongly supershifted Zac-DNA complexes (
). Anti-p300 antibodies (α-p300-N and α-p300-C) were ineffective in the absence of ectopic p300 (lanes 3 and 4) but strongly supershifted Zac-DNA complexes (

 ) upon p300 cotransfection (lanes 7 and 8). Preimmune sera were ineffective (lanes 9 and 10). (C) EMSA. Adjusted amounts of Zac (0.2 μg of pRK7Flag-Zac) and ZacΔC1 (0.04 μg of pRK7Flag-ZacΔC1) were transfected in PA-TU cells and tested as described above.
) upon p300 cotransfection (lanes 7 and 8). Preimmune sera were ineffective (lanes 9 and 10). (C) EMSA. Adjusted amounts of Zac (0.2 μg of pRK7Flag-Zac) and ZacΔC1 (0.04 μg of pRK7Flag-ZacΔC1) were transfected in PA-TU cells and tested as described above.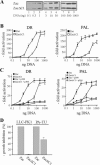

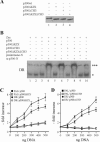
 ) were unaffected by preimmune-N or α-p300-N sera in the absence of p300 (lanes 1 and 2). They were indistinguishably supershifted (
) were unaffected by preimmune-N or α-p300-N sera in the absence of p300 (lanes 1 and 2). They were indistinguishably supershifted (

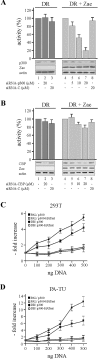
 ) by α-p300-N but not by preimmune-N serum in the presence of p300, p300ΔKIX, or p300ΔCH3 (lanes 3 to 8), while p300ΔKIXΔCH3 was ineffective (lanes 9 and 10). (C and D) Zac (0.1 μg of pRK7Flag-Zac) and DR or PAL reporters (2 μg each) were cotransfected with increasing amounts of p300, p300ΔKIX, and p300ΔCH3 into PA-TU cells. Indicated amounts of DNA refer to p300 and appropriately adjusted amounts of the derivatives.
) by α-p300-N but not by preimmune-N serum in the presence of p300, p300ΔKIX, or p300ΔCH3 (lanes 3 to 8), while p300ΔKIXΔCH3 was ineffective (lanes 9 and 10). (C and D) Zac (0.1 μg of pRK7Flag-Zac) and DR or PAL reporters (2 μg each) were cotransfected with increasing amounts of p300, p300ΔKIX, and p300ΔCH3 into PA-TU cells. Indicated amounts of DNA refer to p300 and appropriately adjusted amounts of the derivatives.


Similar articles
-
Activation of p300 histone acetyltransferase activity and acetylation of the androgen receptor by bombesin in prostate cancer cells.Oncogene. 2006 Mar 30;25(14):2011-21. doi: 10.1038/sj.onc.1209231. Oncogene. 2006. PMID: 16434977
-
Alternative splicing of the imprinted candidate tumor suppressor gene ZAC regulates its antiproliferative and DNA binding activities.Oncogene. 2001 Mar 8;20(10):1246-53. doi: 10.1038/sj.onc.1204237. Oncogene. 2001. PMID: 11313869
-
Enhancement of the p300 HAT activity by HIV-1 Tat on chromatin DNA.Virology. 2001 Oct 25;289(2):312-26. doi: 10.1006/viro.2001.1129. Virology. 2001. PMID: 11689053
-
HATs off to PIC assembly.Mol Cell. 2006 Sep 15;23(6):776-7. doi: 10.1016/j.molcel.2006.08.022. Mol Cell. 2006. PMID: 16973430 Review.
-
Function of the ING family of PHD proteins in cancer.Int J Biochem Cell Biol. 2005 May;37(5):1054-65. doi: 10.1016/j.biocel.2004.09.008. Int J Biochem Cell Biol. 2005. PMID: 15743678 Review.
Cited by
-
C2H2-Type Zinc Finger Proteins: Evolutionarily Old and New Partners of the Nuclear Hormone Receptors.Nucl Recept Signal. 2018 Oct 24;15:1550762918801071. doi: 10.1177/1550762918801071. eCollection 2018. Nucl Recept Signal. 2018. PMID: 30718982 Free PMC article. Review.
-
PLAGL1 gene function during hepatoma cells proliferation.Oncotarget. 2018 Aug 28;9(67):32775-32794. doi: 10.18632/oncotarget.25996. eCollection 2018 Aug 28. Oncotarget. 2018. PMID: 30214684 Free PMC article.
-
Neonatal diabetes mellitus.Endocr Rev. 2008 May;29(3):265-91. doi: 10.1210/er.2007-0029. Epub 2008 Apr 24. Endocr Rev. 2008. PMID: 18436707 Free PMC article. Review.
-
Zac1 regulates cell cycle arrest in neuronal progenitors via Tcf4.Mol Cell Biol. 2014 Mar;34(6):1020-30. doi: 10.1128/MCB.01195-13. Epub 2014 Jan 6. Mol Cell Biol. 2014. PMID: 24396065 Free PMC article.
-
Toward fully exploiting the therapeutic potential of marketed pharmaceuticals: the use of octreotide and chloroquine in oncology.Onco Targets Ther. 2018 Dec 31;12:319-339. doi: 10.2147/OTT.S182685. eCollection 2019. Onco Targets Ther. 2018. PMID: 30643430 Free PMC article. Review.
References
-
- Abdollahi, A., A. K. Godwin, P. D. Miller, L. A. Getts, D. C. Schultz, T. Taguchi, J. R. Testa, and T. C. Hamilton. 1997. Identification of a gene containing zinc-finger motifs based on lost expression in malignantly transformed rat ovarian surface epithelial cells. Cancer Res. 57:2029-2034. - PubMed
-
- Alam, S., D. Zinyk, L. Ma, and C. Schuurmans. 2005. Members of the Plag gene family are expressed in complementary and overlapping regions in the developing murine nervous system. Dev. Dyn. 234:772-782. - PubMed
-
- Arima, T., T. Kamikihara, T. Hayashida, K. Kato, T. Inoue, Y. Shirayoshi, M. Oshimura, H. Soejima, T. Mukai, and N. Wake. 2005. Zac, Lit1 (Kcnq1Ot1) and P57(Kip2) (Cdkn1C) are in an imprinted gene network that may play a role in Beckwith-Wiedemann syndrome. Nucleic Acids Res. 33:2650-2660. - PMC - PubMed
-
- Basyuk, E., V. Coulon, D. A. Le, M. Coisy-Quivy, J. P. Moles, A. Gandarillas, and L. Journot. 2005. The candidate tumor suppressor gene ZAC is involved in keratinocyte differentiation and its expression is lost in basal cell carcinomas. Mol. Cancer Res. 3:483-492. - PubMed
-
- Bilanges, B., A. Varrault, E. Basyuk, C. Rodriguez, A. Mazumdar, C. Pantaloni, J. Bockaert, C. Theillet, D. Spengler, and L. Journot. 1999. Loss of expression of the candidate tumor suppressor gene ZAC in breast cancer cell lines and primary tumors. Oncogene 18:3979-3988. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
