Large nucleotide-dependent movement of the N-terminal domain of the ClpX chaperone
- PMID: 16810315
- PMCID: PMC1523177
- DOI: 10.1038/sj.emboj.7601223
Large nucleotide-dependent movement of the N-terminal domain of the ClpX chaperone
Abstract
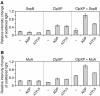
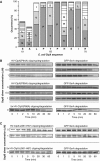
The ClpXP ATPase-protease complex is a major component of the protein quality control machinery in the cell. A ClpX subunit consists of an N-terminal zinc binding domain (ZBD) and a C-terminal AAA+ domain. ClpX oligomerizes into a hexamer with the AAA+ domains forming the base of the hexamer and the ZBDs extending out of the base. Here, we report that ClpX switches between a capture and a feeding conformation. ZBDs in ClpX undergo large nucleotide-dependent block movement towards ClpP and into the AAA+ ring. This motion is modulated by the ClpX cofactor, SspB. Evidence for this movement was initially obtained by the surprising observation that an N-terminal extension on ClpX is clipped by bound ClpP in functional ClpXP complexes. Protease-protection, crosslinking, and light scattering experiments further support these findings.
Figures





References
-
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R (2000) The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature 403: 800–805 - PubMed
-
- Bolon DN, Grant RA, Baker TA, Sauer RT (2004a) Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol Cell 16: 343–350 - PubMed
-
- Bolon DN, Wah DA, Hersch GL, Baker TA, Sauer RT (2004b) Bivalent tethering of SspB to ClpXP is required for efficient substrate delivery: a protein-design study. Mol Cell 13: 443–449 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

