X-ray crystal structure of MENT: evidence for functional loop-sheet polymers in chromatin condensation
- PMID: 16810322
- PMCID: PMC1500978
- DOI: 10.1038/sj.emboj.7601201
X-ray crystal structure of MENT: evidence for functional loop-sheet polymers in chromatin condensation
Abstract
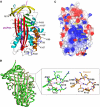
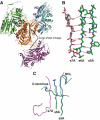
Most serpins are associated with protease inhibition, and their ability to form loop-sheet polymers is linked to conformational disease and the human serpinopathies. Here we describe the structural and functional dissection of how a unique serpin, the non-histone architectural protein, MENT (Myeloid and Erythroid Nuclear Termination stage-specific protein), participates in DNA and chromatin condensation. Our data suggest that MENT contains at least two distinct DNA-binding sites, consistent with its simultaneous binding to the two closely juxtaposed linker DNA segments on a nucleosome. Remarkably, our studies suggest that the reactive centre loop, a region of the MENT molecule essential for chromatin bridging in vivo and in vitro, is able to mediate formation of a loop-sheet oligomer. These data provide mechanistic insight into chromatin compaction by a non-histone architectural protein and suggest how the structural plasticity of serpins has adapted to mediate physiological, rather than pathogenic, loop-sheet linkages.
Figures






References
-
- Carrell RW, Lomas DA (1997) Conformational disease. Lancet 350: 134–138 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

