Evidence of structural genomic region recombination in Hepatitis C virus
- PMID: 16813646
- PMCID: PMC1533811
- DOI: 10.1186/1743-422X-3-53
Evidence of structural genomic region recombination in Hepatitis C virus
Abstract
Background/aim: Hepatitis C virus (HCV) has been the subject of intense research and clinical investigation as its major role in human disease has emerged. Although homologous recombination has been demonstrated in many members of the family Flaviviridae, to which HCV belongs, there have been few studies reporting recombination on natural populations of HCV. Recombination break-points have been identified in non structural proteins of the HCV genome. Given the implications that recombination has for RNA virus evolution, it is clearly important to determine the extent to which recombination plays a role in HCV evolution. In order to gain insight into these matters, we have performed a phylogenetic analysis of 89 full-length HCV strains from all types and sub-types, isolated all over the world, in order to detect possible recombination events.
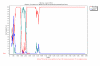
Method: Putative recombinant sequences were identified with the use of SimPlot program. Recombination events were confirmed by bootscaning, using putative recombinant sequence as a query.
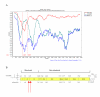
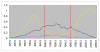
Results: Two crossing over events were identified in the E1/E2 structural region of an intra-typic (1a/1c) recombinant strain.
Conclusion: Only one of 89 full-length strains studied resulted to be a recombinant HCV strain, revealing that homologous recombination does not play an extensive roll in HCV evolution. Nevertheless, this mechanism can not be denied as a source for generating genetic diversity in natural populations of HCV, since a new intra-typic recombinant strain was found. Moreover, the recombination break-points were found in the structural region of the HCV genome.
Figures
References
-
- Chambers TJ, Fan X, Droll DA, Hembrador E, Slater T, Nickells MW, Dustin LB, Dibisceglie AM. Quasispecies heterogeneity within the E1/E2 region as a pretreatment variable during pegylated interferon therapy of chronic hepatitis C virus infection. J Virol. 2005;79:3071–3083. doi: 10.1128/JVI.79.5.3071-3083.2005. - DOI - PMC - PubMed
-
- Laskus T, Wilkinson J, Gallegos-Orozco JF, Radkowski M, Adair DM, Nowicki M, Operskalsi E, Buskell Z, Seeff LB, Vargas H, Rakela J. Analysis of hepatitis C virus quasispecies transmission and evolution in patients infected through blood transfusion. Gastroenterology. 2004;127:764–776. doi: 10.1053/j.gastro.2004.06.005. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

