Bioinformatic discovery of microRNA precursors from human ESTs and introns
- PMID: 16813663
- PMCID: PMC1526439
- DOI: 10.1186/1471-2164-7-164
Bioinformatic discovery of microRNA precursors from human ESTs and introns
Abstract
Background: MicroRNAs (miRNAs) function in many physiological processes, and their discovery is beneficial for further studying their physiological functions. However, many of the miRNAs predicted from genomic sequences have not been experimentally validated to be authentic expressed RNA transcripts, thereby decreasing the reliability of miRNA discovery. To overcome this problem, we examined expressed transcripts - ESTs and intronic sequences - to identify novel miRNAs as well as their target genes.
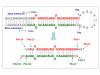
Results: To facilitate our approach, we developed our scanning method using criteria based on the features of 207 known human pre-miRNAs to discriminate miRNAs from random sequences. We identified 208 candidate hairpins in human ESTs and human reference gene intronic sequences, 52 of which are known pre-miRNAs. The discovery pipeline performance was further assessed using 130 newly updated pre-miRNA and randomly selected sequences. We achieved sensitivity of 85% (110/130) and overall specificity of 49.7% using this method. Because miRNAs are evolutionarily conserved regulators of gene expression, it is expected that their host genes and target genes should have respective phylogenetic orthologs. Our results confirmed that, in certain mammals, the host genes carrying the same miRNAs are orthologs, as previously reported. Moreover, this observation is also the case for some of the miRNA target genes.
Conclusion: We have predicted 208 human pre-miRNA candidates and over 10,000 putative human target genes. Using sequence information from ESTs and introns ensures that the predicted pre-miRNA candidates are expressed and the combined expression transcription information from ESTs and introns makes our prediction results more decisive with regard to expressed pre-miRNAs.
Figures

Similar articles
-
Discovering conserved insect microRNAs from expressed sequence tags.J Insect Physiol. 2010 Dec;56(12):1763-9. doi: 10.1016/j.jinsphys.2010.07.007. Epub 2010 Jul 30. J Insect Physiol. 2010. PMID: 20655920
-
Intronic microRNA: discovery and biological implications.DNA Cell Biol. 2007 Apr;26(4):195-207. doi: 10.1089/dna.2006.0558. DNA Cell Biol. 2007. PMID: 17465886
-
Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine.BMC Bioinformatics. 2005 Dec 29;6:310. doi: 10.1186/1471-2105-6-310. BMC Bioinformatics. 2005. PMID: 16381612 Free PMC article.
-
Isolation and identification of gene-specific microRNAs.Methods Mol Biol. 2006;342:313-20. doi: 10.1385/1-59745-123-1:313. Methods Mol Biol. 2006. PMID: 16957385 Review.
-
Computational prediction of microRNA genes.Methods Mol Biol. 2014;1097:437-56. doi: 10.1007/978-1-62703-709-9_20. Methods Mol Biol. 2014. PMID: 24639171 Review.
Cited by
-
MicroRNA Biogenesis, Gene Regulation Mechanisms, and Availability in Foods.Noncoding RNA. 2024 Oct 11;10(5):52. doi: 10.3390/ncrna10050052. Noncoding RNA. 2024. PMID: 39452838 Free PMC article. Review.
-
MicroRNAs as Potential Graft Rejection or Tolerance Biomarkers and Their Dilemma in Clinical Routines Behaving like Devilish, Angelic, or Frightening Elements.Biomedicines. 2024 Jan 5;12(1):116. doi: 10.3390/biomedicines12010116. Biomedicines. 2024. PMID: 38255221 Free PMC article. Review.
-
Differential expression patterns of conserved miRNAs and isomiRs during Atlantic halibut development.BMC Genomics. 2012 Jan 10;13:11. doi: 10.1186/1471-2164-13-11. BMC Genomics. 2012. PMID: 22233483 Free PMC article.
-
Vir-Mir db: prediction of viral microRNA candidate hairpins.Nucleic Acids Res. 2008 Jan;36(Database issue):D184-9. doi: 10.1093/nar/gkm610. Epub 2007 Aug 15. Nucleic Acids Res. 2008. PMID: 17702763 Free PMC article.
-
Origin and evolution of human microRNAs from transposable elements.Genetics. 2007 Jun;176(2):1323-37. doi: 10.1534/genetics.107.072553. Epub 2007 Apr 15. Genetics. 2007. PMID: 17435244 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

