Bacteriorhodopsin chimeras containing the third cytoplasmic loop of bovine rhodopsin activate transducin for GTP/GDP exchange
- PMID: 16815918
- PMCID: PMC2265101
- DOI: 10.1110/ps.062192306
Bacteriorhodopsin chimeras containing the third cytoplasmic loop of bovine rhodopsin activate transducin for GTP/GDP exchange
Abstract
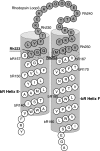
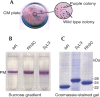
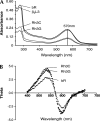
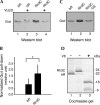
The mechanisms by which G-protein-coupled receptors (GPCRs) activate G-proteins are not well understood due to the lack of atomic structures of GPCRs in an active form or in GPCR/G-protein complexes. For study of GPCR/G-protein interactions, we have generated a series of chimeras by replacing the third cytoplasmic loop of a scaffold protein bacteriorhodopsin (bR) with various lengths of cytoplasmic loop 3 of bovine rhodopsin (Rh), and one such chimera containing loop 3 of the human beta2-adrenergic receptor. The chimeras expressed in the archaeon Halobacterium salinarum formed purple membrane lattices thus facilitating robust protein purification. Retinal was correctly incorporated into the chimeras, as determined by spectrophotometry. A 2D crystal (lattice) was evidenced by circular dichroism analysis, and proper organization of homotrimers formed by the bR/Rh loop 3 chimera Rh3C was clearly illustrated by atomic force microscopy. Most interestingly, Rh3C (and Rh3G to a lesser extent) was functional in activation of GTPgamma35S/GDP exchange of the transducin alpha subunit (Galphat) at a level 3.5-fold higher than the basal exchange. This activation was inhibited by GDP and by a high-affinity peptide analog of the Galphat C terminus, indicating specificity in the exchange reaction. Furthermore, a specific physical interaction between the chimera Rh3C loop 3 and the Galphat C terminus was demonstrated by cocentrifugation of transducin with Rh3C. This Galphat-activating bR/Rh chimera is highly likely to be a useful tool for studying GPCR/G-protein interactions.
Figures



Similar articles
-
Chimeric microbial rhodopsins containing the third cytoplasmic loop of bovine rhodopsin.Biophys J. 2011 Apr 20;100(8):1874-82. doi: 10.1016/j.bpj.2011.02.054. Biophys J. 2011. PMID: 21504723 Free PMC article.
-
Two light-transducing membrane proteins: bacteriorhodopsin and the mammalian rhodopsin.Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1166-71. doi: 10.1073/pnas.90.4.1166. Proc Natl Acad Sci U S A. 1993. PMID: 8433978 Free PMC article.
-
Conformations of the rhodopsin third cytoplasmic loop grafted onto bacteriorhodopsin.Structure. 2000 Jun 15;8(6):643-53. doi: 10.1016/s0969-2126(00)00151-9. Structure. 2000. PMID: 10873864
-
Mechanism of G-protein activation by rhodopsin.Photochem Photobiol. 2007 Jan-Feb;83(1):70-5. doi: 10.1562/2006-03-22-IR-854. Photochem Photobiol. 2007. PMID: 16800722 Review.
-
Complexes between photoactivated rhodopsin and transducin: progress and questions.Biochem J. 2010 Apr 28;428(1):1-10. doi: 10.1042/BJ20100270. Biochem J. 2010. PMID: 20423327 Free PMC article. Review.
Cited by
-
Chimeric microbial rhodopsins containing the third cytoplasmic loop of bovine rhodopsin.Biophys J. 2011 Apr 20;100(8):1874-82. doi: 10.1016/j.bpj.2011.02.054. Biophys J. 2011. PMID: 21504723 Free PMC article.
-
Engineering and Production of the Light-Driven Proton Pump Bacteriorhodopsin in 2D Crystals for Basic Research and Applied Technologies.Methods Protoc. 2020 Jul 22;3(3):51. doi: 10.3390/mps3030051. Methods Protoc. 2020. PMID: 32707904 Free PMC article.
-
Disease-causing mutation in GPR54 reveals the importance of the second intracellular loop for class A G-protein-coupled receptor function.J Biol Chem. 2008 Nov 7;283(45):31068-78. doi: 10.1074/jbc.M805251200. Epub 2008 Sep 4. J Biol Chem. 2008. PMID: 18772143 Free PMC article.
-
Rhodopsins: An Excitingly Versatile Protein Species for Research, Development and Creative Engineering.Front Chem. 2022 Jun 22;10:879609. doi: 10.3389/fchem.2022.879609. eCollection 2022. Front Chem. 2022. PMID: 35815212 Free PMC article. Review.
-
Engineering the catalytic activity of an Antarctic PET-degrading enzyme by loop exchange.Protein Sci. 2023 Sep;32(9):e4757. doi: 10.1002/pro.4757. Protein Sci. 2023. PMID: 37574805 Free PMC article.
References
-
- Abdulaev N.G., Strassmaier T.T., Ngo T., Chen R., Luecke H., Oprian D.D., Ridge K.D. 2002. Grafting segments from the extracellular surface of CCR5 onto a bacteriorhodopsin transmembrane scaffold confers HIV-1 coreceptor activity. Structure 10 515–525. - PubMed
-
- Ahl P.L., Price R., Smuda J., Gaber B.P., Singh A. 1990. Insertion of bacteriorhodopsin into polymerized diacetylenic phosphatidylcholine bilayers. Biochim. Biophys. Acta 1028 141–153. - PubMed
-
- Ahumada A., Slusarski D.C., Liu X., Moon R.T., Malbon C.C., Wang H.Y. 2002. Signaling of rat Frizzled-2 through phosphodiesterase and cyclic GMP. Science 298 2006–2010. - PubMed
-
- Altenbach C., Yang K., Farrens D.L., Farahbakhsh Z.T., Khorana H.G., Hubbell W.L. 1996. Structural features and light-dependent changes in the cytoplasmic interhelical E-F loop region of rhodopsin: A site-directed spin-labeling study. Biochemistry 35 12470–12478. - PubMed
-
- Brith-Linder M. and Rosenheck K. 1977. The circular dichroism of bacteriorhodopsin: Asymmetry and light-scattering distortions. FEBS Lett. 76 41–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

