A limited universe of membrane protein families and folds
- PMID: 16815920
- PMCID: PMC2242558
- DOI: 10.1110/ps.062109706
A limited universe of membrane protein families and folds
Abstract
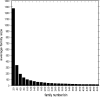
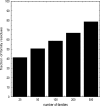
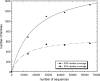
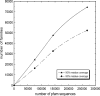
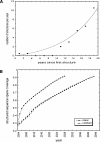
One of the goals of structural genomics is to obtain a structural representative of almost every fold in nature. A recent estimate suggests that 70%-80% of soluble protein domains identified in the first 1000 genome sequences should be covered by about 25,000 structures-a reasonably achievable goal. As no current estimates exist for the number of membrane protein families, however, it is not possible to know whether family coverage is a realistic goal for membrane proteins. Here we find that virtually all polytopic helical membrane protein families are present in the already known sequences so we can make an estimate of the total number of families. We find that only approximately 700 polytopic membrane protein families account for 80% of structured residues and approximately 1700 cover 90% of structured residues. While apparently a finite and reachable goal, we estimate that it will likely take more than three decades to obtain the structures needed for 90% residue coverage, if current trends continue.
Figures

Similar articles
-
Defining the fold space of membrane proteins: the CAMPS database.Proteins. 2006 Sep 1;64(4):906-22. doi: 10.1002/prot.21081. Proteins. 2006. PMID: 16802318
-
Protein family clustering for structural genomics.J Mol Biol. 2005 Oct 28;353(3):744-59. doi: 10.1016/j.jmb.2005.08.058. Epub 2005 Sep 9. J Mol Biol. 2005. PMID: 16185712
-
Structural imperatives impose diverse evolutionary constraints on helical membrane proteins.Proc Natl Acad Sci U S A. 2009 Oct 20;106(42):17747-50. doi: 10.1073/pnas.0906390106. Epub 2009 Oct 6. Proc Natl Acad Sci U S A. 2009. PMID: 19815527 Free PMC article.
-
Folding energetics and oligomerization of polytopic α-helical transmembrane proteins.Arch Biochem Biophys. 2014 Dec 15;564:281-96. doi: 10.1016/j.abb.2014.07.017. Epub 2014 Jul 21. Arch Biochem Biophys. 2014. PMID: 25057769 Review.
-
Folding and Insertion of Transmembrane Helices at the ER.Int J Mol Sci. 2021 Nov 26;22(23):12778. doi: 10.3390/ijms222312778. Int J Mol Sci. 2021. PMID: 34884581 Free PMC article. Review.
Cited by
-
Building a virtual ligand screening pipeline using free software: a survey.Brief Bioinform. 2016 Mar;17(2):352-66. doi: 10.1093/bib/bbv037. Epub 2015 Jun 20. Brief Bioinform. 2016. PMID: 26094053 Free PMC article. Review.
-
Phylogeny of the Vitamin K 2,3-Epoxide Reductase (VKOR) Family and Evolutionary Relationship to the Disulfide Bond Formation Protein B (DsbB) Family.Nutrients. 2015 Jul 29;7(8):6224-49. doi: 10.3390/nu7085281. Nutrients. 2015. PMID: 26230708 Free PMC article.
-
Folding and Misfolding of Human Membrane Proteins in Health and Disease: From Single Molecules to Cellular Proteostasis.Chem Rev. 2019 May 8;119(9):5537-5606. doi: 10.1021/acs.chemrev.8b00532. Epub 2019 Jan 4. Chem Rev. 2019. PMID: 30608666 Free PMC article.
-
A survey of integral alpha-helical membrane proteins.J Struct Funct Genomics. 2009 Dec;10(4):269-80. doi: 10.1007/s10969-009-9069-8. Epub 2009 Sep 17. J Struct Funct Genomics. 2009. PMID: 19760129 Free PMC article.
-
Structural and functional analysis of transmembrane segment IV of the salt tolerance protein Sod2.J Biol Chem. 2013 Aug 23;288(34):24609-24. doi: 10.1074/jbc.M113.483065. Epub 2013 Jul 8. J Biol Chem. 2013. PMID: 23836910 Free PMC article.
References
-
- Bass R.B., Strop P., Barclay M., Rees D.C. 2002. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science 298: 1582–1587. - PubMed
-
- Berry E.A., Guergova-Kuras M., Huang L.S., Crofts A.R. 2000. Structure and function of cytochrome bc complexes. Annu. Rev. Biochem. 69: 1005–1075. - PubMed
-
- Bowie J.U. 1997a. Helix packing angle preferences. Nat. Struct. Biol. 4: 915–917. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

