mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes
- PMID: 16815998
- PMCID: PMC1522082
- DOI: 10.1101/gad.1424106
mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes
Abstract
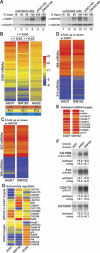
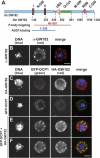
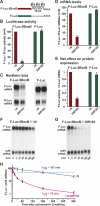
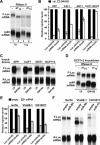
MicroRNAs (miRNAs) silence the expression of target genes post-transcriptionally. Their function is mediated by the Argonaute proteins (AGOs), which colocalize to P-bodies with mRNA degradation enzymes. Mammalian P-bodies are also marked by the GW182 protein, which interacts with the AGOs and is required for miRNA function. We show that depletion of GW182 leads to changes in mRNA expression profiles strikingly similar to those observed in cells depleted of the essential Drosophila miRNA effector AGO1, indicating that GW182 functions in the miRNA pathway. When GW182 is bound to a reporter transcript, it silences its expression, bypassing the requirement for AGO1. Silencing by GW182 is effected by changes in protein expression and mRNA stability. Similarly, miRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay, and both mechanisms require GW182. mRNA degradation, but not translational repression, by GW182 or miRNAs is inhibited in cells depleted of CAF1, NOT1, or the decapping DCP1:DCP2 complex. We further show that the N-terminal GW repeats of GW182 interact with the PIWI domain of AGO1. Our findings indicate that GW182 links the miRNA pathway to mRNA degradation by interacting with AGO1 and promoting decay of at least a subset of miRNA targets.
Figures

References
-
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
-
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Ding L., Spencer A., Morita K., Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell. 2005;19:437–447. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
