A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus
- PMID: 16816137
- PMCID: PMC1533990
- DOI: 10.1105/tpc.105.039966
A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus
Abstract
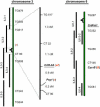
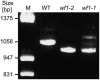
Carotenoids and their oxygenated derivatives xanthophylls play essential roles in the pigmentation of flowers and fruits. Wild-type tomato (Solanum lycopersicum) flowers are intensely yellow due to accumulation of the xanthophylls neoxanthin and violaxanthin. To study the regulation of xanthophyll biosynthesis, we analyzed the mutant white-flower (wf). It was found that the recessive wf phenotype is caused by mutations in a flower-specific beta-ring carotene hyroxylase gene (CrtR-b2). Two deletions and one exon-skipping mutation in different CrtR-b2 wf alleles abolish carotenoid biosynthesis in flowers but not leaves, where the homologous CrtR-b1 is constitutively expressed. A second beta-carotene hydroxylase enzyme as well as flower- and fruit-specific geranylgeranyl diphosphate synthase, phytoene synthase, and lycopene beta-cyclase together define a carotenoid biosynthesis pathway active in chromoplasts only, underscoring the crucial role of gene duplication in specialized plant metabolic pathways. We hypothesize that this pathway in tomato was initially selected during evolution to enhance flower coloration and only later recruited to enhance fruit pigmentation. The elimination of beta-carotene hydroxylation in wf petals results in an 80% reduction in total carotenoid concentration, possibly caused by the inability of petals to store high concentrations of carotenoids other than xanthophylls and by degradation of beta-carotene, which accumulates as a result of the wf mutation but is not due to altered expression of genes in the biosynthetic pathway.
Figures








References
-
- Auldridge, M.E., Block, A., Vogel, J.T., Dabney-Smith, C., Mila, I., Bouzayen, M., Magallanes-Lundback, M., DellaPenna, D., McCarty, D.R., and Klee, H.J. (2006). Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 45 982–993. - PubMed
-
- Bartley, G.E., and Scolnik, P.A. (1993). cDNA cloning, expression during development, and genome mapping of PSY2, a second tomato gene encoding phytoene synthase. J. Biol. Chem. 268 25718–25721. - PubMed
-
- Bartley, G.E., Viitanen, P.V., Bacot, K.O., and Scolnik, P.A. (1992). A tomato gene expressed during fruit ripening encodes an enzyme of the carotenoid biosynthesis pathway. J. Biol. Chem. 267 5036–5039. - PubMed
-
- Bouvier, F., Keller, Y., d'Harlingue, A., and Camara, B. (1998). Xanthophyll biosynthesis: Molecular and functional characterization of carotenoid hydroxylases from pepper fruits (Capsicum annuum L.). Biochim. Biophys. Acta 1391 320–328. - PubMed
-
- Bramley, P.M. (2002). Regulation of carotenoid formation during tomato fruit ripening and development. J. Exp. Bot. 53 2107–2113. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

