GlcNAc-6P levels modulate the expression of Curli fibers by Escherichia coli
- PMID: 16816193
- PMCID: PMC1539958
- DOI: 10.1128/JB.00234-06
GlcNAc-6P levels modulate the expression of Curli fibers by Escherichia coli
Abstract
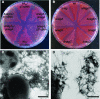
Curli are extracellular surface fibers that are produced by many members of the Enterobacteriaceae and contribute to biofilm formation. The environmental cues that promote biofilm formation are poorly understood. We found that deletion of the N-acetylglucosamine-6-phosphate (GlcNAc-6P) deacetylase gene, nagA, resulted in decreased transcription from the curli-specific promoters csgBA and csgDEFG and a corresponding decrease in curli production in Escherichia coli. nagA is in an operon that contains nagB, nagC, nagD, and nagE, whose products are required for utilization of GlcNAc as a carbon source. NagC is a repressor of the nagBACD and nagE genes in the absence of intracellular GlcNAc-6P. We found that nagC mutants were also defective in curli production. Growth of a wild-type strain on media containing additional GlcNAc reduced curli gene transcription to a level similar to the level observed when nagA was deleted. The defect in curli production in nagA or nagC mutants was alleviated by deletion of the GlcNAc transporter gene, nagE. Curli-producing DeltanagA suppressor mutants whose cells were unable to take up GlcNAc were isolated. These results suggest that elevated levels of intracellular GlcNAc-6P signal cells to down-regulate curli gene expression.
Figures
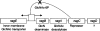



References
-
- Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. - PubMed
-
- Arnqvist, A., A. Olsen, and S. Normark. 1994. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13:1021-1032. - PubMed
-
- Bian, Z., A. Brauner, Y. Li, and S. Normark. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 181:602-612. - PubMed
-
- Bian, Z., Z. Q. Yan, G. K. Hansson, P. Thoren, and S. Normark. 2001. Activation of inducible nitric oxide synthase/nitric oxide by curli fibers leads to a fall in blood pressure during systemic Escherichia coli infection in mice. J. Infect. Dis. 183:612-619. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

