Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides
- PMID: 16816200
- PMCID: PMC1539962
- DOI: 10.1128/JB.00300-06
Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides
Abstract
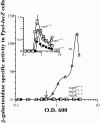
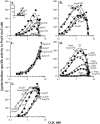
In Bacillus subtilis, extracellular peptide signaling regulates several biological processes. Secreted Phr signaling peptides are imported into the cell and act intracellularly to antagonize the activity of regulators known as Rap proteins. B. subtilis encodes several Rap proteins and Phr peptides, and the processes regulated by many of these Rap proteins and Phr peptides are unknown. We used DNA microarrays to characterize the roles that several rap-phr signaling modules play in regulating gene expression. We found that rapK-phrK regulates the expression of a number of genes activated by the response regulator ComA. ComA activates expression of genes involved in competence development and the production of several secreted products. Two Phr peptides, PhrC and PhrF, were previously known to stimulate the activity of ComA. We assayed the roles that PhrC, PhrF, and PhrK play in regulating gene expression and found that these three peptides stimulate ComA-dependent gene expression to different levels and are all required for full expression of genes activated by ComA. The involvement of multiple Rap proteins and Phr peptides allows multiple physiological cues to be integrated into a regulatory network that modulates the timing and magnitude of the ComA response.
Figures


References
-
- Ansaldi, M., D. Marolt, T. Stebe, I. Mandic-Mulec, and D. Dubnau. 2002. Specific activation of the Bacillus quorum-sensing systems by isoprenylated pheromone variants. Mol. Microbiol. 44:1561-1573. - PubMed
-
- Berka, R. M., J. Hahn, M. Albano, I. Draskovic, M. Persuh, X. Cui, A. Sloma, W. Widner, and D. Dubnau. 2002. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43:1331-1345. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

