Regulation of the Helicobacter pylori Fe-S cluster synthesis protein NifS by iron, oxidative stress conditions, and fur
- PMID: 16816209
- PMCID: PMC1539971
- DOI: 10.1128/JB.00104-06
Regulation of the Helicobacter pylori Fe-S cluster synthesis protein NifS by iron, oxidative stress conditions, and fur
Abstract
Transcription of both chromosomal and extrachromosomally introduced nifS was regulated (up-expressed) by oxygen or by supplemental iron conditions. This up-expression was not observed in a fur mutant strain background or when an iron chelator was added. Iron-bound Fur (but not apo-Fur) recognized the nifS promoter, and Fur bound significantly farther upstream (-155 bp to -190 bp and -210 to -240 bp) in the promoter than documented Helicobacter pylori Fur binding regions. This binding was stronger than Fur recognition of the flgE or napA promoter and includes a Fur recognition sequence common to the H. pylori pfr and sodB upstream areas. Studies of Fur-regulated genes in H. pylori have indicated that apo-Fur acts as a repressor, but our results demonstrate that iron-bound Fur activates (nifS) transcription.
Figures
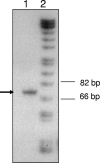
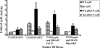
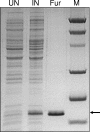
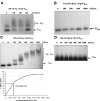


References
-
- Alamuri, P., and R. J. Maier. 2004. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:1397-1406. - PubMed
-
- Blaser, M. J. 1997. The versatility of Helicobacter pylori in the adaptation to the human stomach. J. Physiol. Pharmacol. 48:307-314. - PubMed
-
- Cooksley, C., P. J. Jenks, A. Green, A. Cockayne, R. P. Logan, and K. R. Hardie. 2003. NapA protects Helicobacter pylori from oxidative stress damage, and its production is influenced by the ferric uptake regulator. J. Med. Microbiol. 52:461-469. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

