Using ESTs to improve the accuracy of de novo gene prediction
- PMID: 16817966
- PMCID: PMC1534067
- DOI: 10.1186/1471-2105-7-327
Using ESTs to improve the accuracy of de novo gene prediction
Abstract
Background: ESTs are a tremendous resource for determining the exon-intron structures of genes, but even extensive EST sequencing tends to leave many exons and genes untouched. Gene prediction systems based exclusively on EST alignments miss these exons and genes, leading to poor sensitivity. De novo gene prediction systems, which ignore ESTs in favor of genomic sequence, can predict such "untouched" exons, but they are less accurate when predicting exons to which ESTs align. TWINSCAN is the most accurate de novo gene finder available for nematodes and N-SCAN is the most accurate for mammals, as measured by exact CDS gene prediction and exact exon prediction.
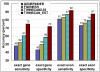
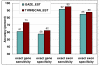
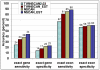
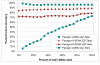
Results: TWINSCAN_EST is a new system that successfully combines EST alignments with TWINSCAN. On the whole C. elegans genome TWINSCAN_EST shows 14% improvement in sensitivity and 13% in specificity in predicting exact gene structures compared to TWINSCAN without EST alignments. Not only are the structures revealed by EST alignments predicted correctly, but these also constrain the predictions without alignments, improving their accuracy. For the human genome, we used the same approach with N-SCAN, creating N-SCAN_EST. On the whole genome, N-SCAN_EST produced a 6% improvement in sensitivity and 1% in specificity of exact gene structure predictions compared to N-SCAN.
Conclusion: TWINSCAN_EST and N-SCAN_EST are more accurate than TWINSCAN and N-SCAN, while retaining their ability to discover novel genes to which no ESTs align. Thus, we recommend using the EST versions of these programs to annotate any genome for which EST information is available.TWINSCAN_EST and N-SCAN_EST are part of the TWINSCAN open source software package http://genes.cse.wustl.edu/distribution/download_TS.html.
Figures
References
-
- Guigó R, Dermitzakis ET, Agarwal P, Ponting C, Parra G, Reymond A, Abril JF, Keibler E, Lyle R, Ucla C, Antonarakis SE, Brent MR. Comparison of mouse and human genomes followed by experimental verification yields an estimated 1,019 additional genes. Proc Natl Acad Sci U S A. 2003;100:1140–1145. doi: 10.1073/pnas.0337561100. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

