Maternal BRG1 regulates zygotic genome activation in the mouse
- PMID: 16818606
- PMCID: PMC1522071
- DOI: 10.1101/gad.1435106
Maternal BRG1 regulates zygotic genome activation in the mouse
Abstract
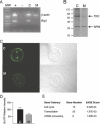
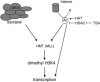
Zygotic genome activation (ZGA) is a nuclear reprogramming event that transforms the genome from transcriptional quiescence at fertilization to robust transcriptional activity shortly thereafter. The ensuing gene expression profile in the cleavage-stage embryo establishes totipotency and is required for further development. Although little is known about the molecular basis of ZGA, oocyte-derived mRNAs and proteins that alter chromatin structure are likely crucial. To test this hypothesis, we generated a maternal-effect mutation of Brg1, which encodes a catalytic subunit of SWI/SNF-related complexes, utilizing Cre-loxP gene targeting. In conditional-mutant females, BRG1-depleted oocytes completed meiosis and were fertilized. However, embryos conceived from BRG1-depleted eggs exhibited a ZGA phenotype including two-cell arrest and reduced transcription for approximately 30% of expressed genes. Genes involved in transcription, RNA processing, and cell cycle regulation were particularly affected. The early embryonic arrest is not a consequence of a defective oocyte because depleting maternal BRG1 after oocyte development is complete by RNA interference (RNAi) also resulted in two-cell arrest. To our knowledge, Brg1 is the first gene required for ZGA in mammals. Depletion of maternal BRG1 did not affect global levels of histone acetylation, whereas dimethyl-H3K4 levels were reduced. These data provide a framework for understanding the mechanism of ZGA.
Figures




References
-
- Agalioti T., Chen G., Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. - PubMed
-
- Aoki F., Worrad D.M., Schultz R.M. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 1997;181:296–307. - PubMed
-
- Ayton P., Sneddon S.F., Palmer D.B., Rosewell I.R., Owen M.J., Young B., Presley R., Subramanian V. Truncation of the Mll gene in exon 5 by gene targeting leads to early preimplantation lethality of homozygous embryos. Genesis. 2001;30:201–212. - PubMed
-
- Bellotto M., Bopp D., Senti K.A., Burke R., Deak P., Maroy P., Dickson B., Basler K., Hafen E. Maternal-effect loci involved in Drosophila oogenesis and embryogenesis: P element-induced mutations on the third chromosome. Int. J. Dev. Biol. 2002;46:149–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
