Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress
- PMID: 16818608
- PMCID: PMC1522074
- DOI: 10.1101/gad.1428206
Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress
Abstract
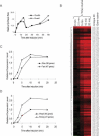
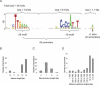
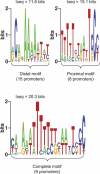
The heat-shock response (HSR), a universal cellular response to heat, is crucial for cellular adaptation. In Escherichia coli, the HSR is mediated by the alternative sigma factor, sigma32. To determine its role, we used genome-wide expression analysis and promoter validation to identify genes directly regulated by sigma32 and screened ORF overexpression libraries to identify sigma32 inducers. We triple the number of genes validated to be transcribed by sigma32 and provide new insights into the cellular role of this response. Our work indicates that the response is propagated as the regulon encodes numerous global transcriptional regulators, reveals that sigma70 holoenzyme initiates from 12% of sigma32 promoters, which has important implications for global transcriptional wiring, and identifies a new role for the response in protein homeostasis, that of protecting complex proteins. Finally, this study suggests that the response protects the cell membrane and responds to its status: Fully 25% of sigma32 regulon members reside in the membrane and alter its functionality; moreover, a disproportionate fraction of overexpressed proteins that induce the response are membrane localized. The intimate connection of the response to the membrane rationalizes why a major regulator of the response resides in that cellular compartment.
Figures

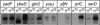
References
-
- Bader M., Muse W., Ballou D.P., Gassner C., Bardwell J.C. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. - PubMed
-
- Beckmann R.P., Mizzen L.E., Welch W.J. Interaction of Hsp 70 with newly synthesized proteins: Implications for protein folding and assembly. Science. 1990;248:850–854. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
