CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells
- PMID: 16818678
- PMCID: PMC2118339
- DOI: 10.1084/jem.20060772
CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells
Abstract
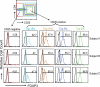
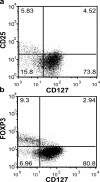
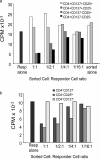
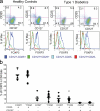
Regulatory T (T reg) cells are critical regulators of immune tolerance. Most T reg cells are defined based on expression of CD4, CD25, and the transcription factor, FoxP3. However, these markers have proven problematic for uniquely defining this specialized T cell subset in humans. We found that the IL-7 receptor (CD127) is down-regulated on a subset of CD4(+) T cells in peripheral blood. We demonstrate that the majority of these cells are FoxP3(+), including those that express low levels or no CD25. A combination of CD4, CD25, and CD127 resulted in a highly purified population of T reg cells accounting for significantly more cells that previously identified based on other cell surface markers. These cells were highly suppressive in functional suppressor assays. In fact, cells separated based solely on CD4 and CD127 expression were anergic and, although representing at least three times the number of cells (including both CD25(+)CD4(+) and CD25(-)CD4(+) T cell subsets), were as suppressive as the "classic" CD4(+)CD25(hi) T reg cell subset. Finally, we show that CD127 can be used to quantitate T reg cell subsets in individuals with type 1 diabetes supporting the use of CD127 as a biomarker for human T reg cells.
Figures



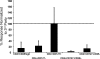
References
-
- Sakaguchi, S., N. Sakaguchi, J. Shimizu, S. Yamazaki, T. Sakihama, M. Itoh, Y. Kuniyasu, T. Nomura, M. Toda, and T. Takahashi. 2001. Immunologic tolerance maintained by CD25+ CD4+ regulatory T cells: their common role in controlling autoimmunity, tumor immunity, and transplantation tolerance. Immunol. Rev. 182:18–32. - PubMed
-
- Chatenoud, L., B. Salomon, and J.A. Bluestone. 2001. Suppressor T cells–they're back and critical for regulation of autoimmunity! Immunol. Rev. 182:149–163. - PubMed
-
- Wood, K.J., and S. Sakaguchi. 2003. Regulatory T cells in transplantation tolerance. Nat. Rev. Immunol. 3:199–210. - PubMed
-
- Singh, B., S. Read, C. Asseman, V. Malmstrom, C. Mottet, L.A. Stephens, R. Stepankova, H. Tlaskalova, and F. Powrie. 2001. Control of intestinal inflammation by regulatory T cells. Immunol. Rev. 182:190–200. - PubMed
-
- Curotto de Lafaille, M.A., and J.J. Lafaille. 2002. CD4(+) regulatory T cells in autoimmunity and allergy. Curr. Opin. Immunol. 14:771–778. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

