Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence
- PMID: 16818717
- PMCID: PMC2064156
- DOI: 10.1083/jcb.200603083
Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence
Abstract
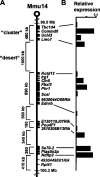
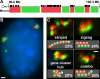
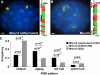
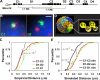
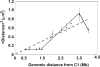
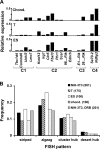
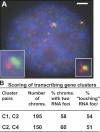
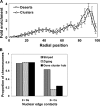
Specific mammalian genes functionally and dynamically associate together within the nucleus. Yet, how an array of many genes along the chromosome sequence can be spatially organized and folded together is unknown. We investigated the 3D structure of a well-annotated, highly conserved 4.3-Mb region on mouse chromosome 14 that contains four clusters of genes separated by gene "deserts." In nuclei, this region forms multiple, nonrandom "higher order" structures. These structures are based on the gene distribution pattern in primary sequence and are marked by preferential associations among multiple gene clusters. Associating gene clusters represent expressed chromatin, but their aggregation is not simply dependent on ongoing transcription. In chromosomes with aggregated gene clusters, gene deserts preferentially align with the nuclear periphery, providing evidence for chromosomal region architecture by specific associations with functional nuclear domains. Together, these data suggest dynamic, probabilistic 3D folding states for a contiguous megabase-scale chromosomal region, supporting the diverse activities of multiple genes and their conserved primary sequence organization.
Figures

Similar articles
-
Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH.J Cell Biol. 2002 Dec 9;159(5):753-63. doi: 10.1083/jcb.200207115. Epub 2002 Dec 9. J Cell Biol. 2002. PMID: 12473685 Free PMC article.
-
Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C).Nat Genet. 2006 Nov;38(11):1348-54. doi: 10.1038/ng1896. Epub 2006 Oct 8. Nat Genet. 2006. PMID: 17033623
-
Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture.Crit Rev Eukaryot Gene Expr. 2000;10(2):179-212. Crit Rev Eukaryot Gene Expr. 2000. PMID: 11186332 Review.
-
Inter- and intra-specific gene-density-correlated radial chromosome territory arrangements are conserved in Old World monkeys.Cytogenet Genome Res. 2005;108(1-3):255-61. doi: 10.1159/000080824. Cytogenet Genome Res. 2005. PMID: 15545738
-
The 4D Nucleome: Genome Compartmentalization in an Evolutionary Context.Biochemistry (Mosc). 2018 Apr;83(4):313-325. doi: 10.1134/S000629791804003X. Biochemistry (Mosc). 2018. PMID: 29626919 Review.
Cited by
-
Chromatin higher-order structure and dynamics.Cold Spring Harb Perspect Biol. 2010 May;2(5):a000596. doi: 10.1101/cshperspect.a000596. Epub 2010 Apr 7. Cold Spring Harb Perspect Biol. 2010. PMID: 20452954 Free PMC article. Review.
-
Dynamic reorganization of the genome shapes the recombination landscape in meiotic prophase.Nat Struct Mol Biol. 2019 Mar;26(3):164-174. doi: 10.1038/s41594-019-0187-0. Epub 2019 Feb 18. Nat Struct Mol Biol. 2019. PMID: 30778236 Free PMC article.
-
Cycles in spatial and temporal chromosomal organization driven by the circadian clock.Nat Struct Mol Biol. 2013 Oct;20(10):1206-13. doi: 10.1038/nsmb.2667. Epub 2013 Sep 22. Nat Struct Mol Biol. 2013. PMID: 24056944 Free PMC article.
-
Combining FISH with localisation microscopy: Super-resolution imaging of nuclear genome nanostructures.Chromosome Res. 2011 Jan;19(1):5-23. doi: 10.1007/s10577-010-9171-6. Chromosome Res. 2011. PMID: 21190132
-
The Self-Organizing Genome: Principles of Genome Architecture and Function.Cell. 2020 Oct 1;183(1):28-45. doi: 10.1016/j.cell.2020.09.014. Epub 2020 Sep 24. Cell. 2020. PMID: 32976797 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

