Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures
- PMID: 16818721
- PMCID: PMC2064167
- DOI: 10.1083/jcb.200604072
Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures
Abstract
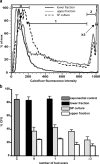
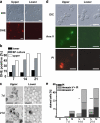
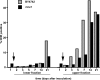
Quiescence is the most common and, arguably, most poorly understood cell cycle state. This is in part because pure populations of quiescent cells are typically difficult to isolate. We report the isolation and characterization of quiescent and nonquiescent cells from stationary-phase (SP) yeast cultures by density-gradient centrifugation. Quiescent cells are dense, unbudded daughter cells formed after glucose exhaustion. They synchronously reenter the mitotic cell cycle, suggesting that they are in a G(0) state. Nonquiescent cells are less dense, heterogeneous, and composed of replicatively older, asynchronous cells that rapidly lose the ability to reproduce. Microscopic and flow cytometric analysis revealed that nonquiescent cells accumulate more reactive oxygen species than quiescent cells, and over 21 d, about half exhibit signs of apoptosis and necrosis. The ability to isolate both quiescent and nonquiescent yeast cells from SP cultures provides a novel, tractable experimental system for studies of quiescence, chronological and replicative aging, apoptosis, and the cell cycle.
Figures

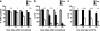
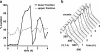
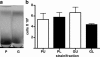

References
-
- Alcasabas, A.A., A.J. Osborn, J. Bachant, F.H. Hu, P.J.H. Werler, K. Bousset, K. Furuya, J.F.X. Diffley, A.M. Carr, and S.J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958–965. - PubMed
-
- Alexander, B.D., and J.R. Perfect. 1997. Antifungal resistance trends towards the year 2000. Implications for therapy and new approaches. Drugs. 54:657–678. - PubMed
-
- Boddy, M.N., P.H.L. Gaillard, W.H. McDonald, P. Shanahan, J.R. Yates, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 107:537–548. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

