The genome of the square archaeon Haloquadratum walsbyi : life at the limits of water activity
- PMID: 16820047
- PMCID: PMC1544339
- DOI: 10.1186/1471-2164-7-169
The genome of the square archaeon Haloquadratum walsbyi : life at the limits of water activity
Abstract
Background: The square halophilic archaeon Haloquadratum walsbyi dominates NaCl-saturated and MgCl2 enriched aquatic ecosystems, which imposes a serious desiccation stress, caused by the extremely low water activity. The genome sequence was analyzed and physiological and physical experiments were carried out in order to reveal how H. walsbyi has specialized into its narrow and hostile ecological niche and found ways to cope with the desiccation stress.
Results: A rich repertoire of proteins involved in phosphate metabolism, phototrophic growth and extracellular protective polymers, including the largest archaeal protein (9159 amino acids), a homolog to eukaryotic mucins, are amongst the most outstanding features. A relatively low GC content (47.9%), 15-20% less than in other halophilic archaea, and one of the lowest coding densities (76.5%) known for prokaryotes might be an indication for the specialization in its unique environment
Conclusion: Although no direct genetic indication was found that can explain how this peculiar organism retains its square shape, the genome revealed several unique adaptive traits that allow this organism to thrive in its specific and extreme niche.
Figures

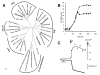

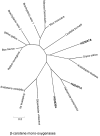

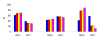
References
-
- Walsby AE. A Square Bacterium. Nature. 1980;283:69–71. doi: 10.1038/283069a0. - DOI
-
- Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML. Cultivation of Walsby's square haloarchaeon. FEMS Microbiol Lett. 2004;238:469–473. - PubMed
-
- Galinski EA, Truper HG. Microbial behaviour in salt-stressed ecosystems. FEMS Microbiology Reviews. 1994;15:95–108. doi: 10.1111/j.1574-6976.1994.tb00128.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

