Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea
- PMID: 16820452
- PMCID: PMC1489360
- DOI: 10.1128/AEM.02919-05
Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea
Abstract
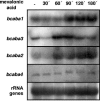
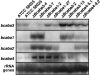
Like several other phytopathogenic fungi, the ascomycete Botrytis cinerea is known to produce the plant hormone abscisic acid (ABA) in axenic culture. Recently, bcaba1, the first fungal gene involved in ABA biosynthesis, was identified. Neighborhood analysis of bcaba1 revealed three further candidate genes of this pathway: a putative P450 monooxygenase-encoding gene (bcaba2), an open reading frame without significant similarities (bcaba3), and a gene probably coding for a short-chain dehydrogenase/reductase (bcaba4). Targeted inactivation of the genes proved the involvement of BcABA2 and BcABA3 in ABA biosynthesis and suggested a contribution of BcABA4. The close linkage of at least three ABA biosynthetic genes is strong evidence for the presence of an abscisic acid gene cluster in B. cinerea.
Figures
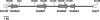


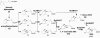
Similar articles
-
Cys2His2 Zinc Finger Transcription Factor BcabaR1 Positively Regulates Abscisic Acid Production in Botrytis cinerea.Appl Environ Microbiol. 2018 Aug 17;84(17):e00920-18. doi: 10.1128/AEM.00920-18. Print 2018 Sep 1. Appl Environ Microbiol. 2018. PMID: 29959241 Free PMC article.
-
Sequencing and transcriptional analysis of the biosynthesis gene cluster of abscisic acid-producing Botrytis cinerea.Int J Mol Sci. 2014 Sep 29;15(10):17396-410. doi: 10.3390/ijms151017396. Int J Mol Sci. 2014. PMID: 25268614 Free PMC article.
-
The P450 monooxygenase BcABA1 is essential for abscisic acid biosynthesis in Botrytis cinerea.Appl Environ Microbiol. 2004 Jul;70(7):3868-76. doi: 10.1128/AEM.70.7.3868-3876.2004. Appl Environ Microbiol. 2004. PMID: 15240257 Free PMC article.
-
Abscisic Acid and Its Producer Botrytis cinerea: Recent Advances and Future Perspectives.J Agric Food Chem. 2025 Aug 6;73(31):19204-19221. doi: 10.1021/acs.jafc.5c05794. Epub 2025 Jul 15. J Agric Food Chem. 2025. PMID: 40662379 Review.
-
Recent Advances in the Study of the Plant Pathogenic Fungus Botrytis cinerea and its Interaction with the Environment.Curr Protein Pept Sci. 2017;18(10):976-989. doi: 10.2174/1389203717666160809160915. Curr Protein Pept Sci. 2017. PMID: 27526927 Review.
Cited by
-
A fungal sesquiterpene biosynthesis gene cluster critical for mutualist-pathogen transition in Colletotrichum tofieldiae.Nat Commun. 2023 Sep 6;14(1):5288. doi: 10.1038/s41467-023-40867-w. Nat Commun. 2023. PMID: 37673872 Free PMC article.
-
Molecular insights into a distinct class of terpenoid cyclases.Nat Commun. 2025 Jan 2;16(1):207. doi: 10.1038/s41467-024-55717-6. Nat Commun. 2025. PMID: 39747870 Free PMC article.
-
Rose WRKY13 promotes disease protection to Botrytis by enhancing cytokinin content and reducing abscisic acid signaling.Plant Physiol. 2023 Jan 2;191(1):679-693. doi: 10.1093/plphys/kiac495. Plant Physiol. 2023. PMID: 36271872 Free PMC article.
-
Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea.PLoS Genet. 2011 Aug;7(8):e1002230. doi: 10.1371/journal.pgen.1002230. Epub 2011 Aug 18. PLoS Genet. 2011. PMID: 21876677 Free PMC article.
-
Abscisic acid deficiency caused by phytoene desaturase silencing is associated with dwarfing syndrome in citrus.Plant Cell Rep. 2019 Aug;38(8):965-980. doi: 10.1007/s00299-019-02418-w. Epub 2019 May 4. Plant Cell Rep. 2019. PMID: 31055623
References
-
- Alexander, N. J., S. P. McCormick, T. M. Larson, and J. E. Jurgenson. 2004. Expression of Tri15 in Fusarium sporotrichioides. Curr. Genet. 45:157-162. - PubMed
-
- Al-Nimri, L. F., and R. C. Coolbaugh. 1991. Conversion of 1′-deoxy-2H-ABA to 2H-ABA in cell-free extracts from Cercospora rosicola. J. Plant Growth Regul. 10:63-66.
-
- Bhatnagar, D., K. C. Ehrlich, and T. E. Cleveland. 2003. Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 61:83-93. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

