Induction of multiple prophages from a marine bacterium: a genomic approach
- PMID: 16820498
- PMCID: PMC1489376
- DOI: 10.1128/AEM.00056-06
Induction of multiple prophages from a marine bacterium: a genomic approach
Abstract
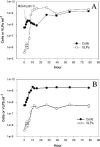
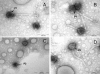
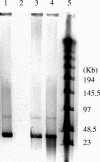
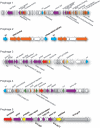
Approximately 70% of sequenced bacterial genomes contain prophage-like structures, yet little effort has been made to use this information to determine the functions of these elements. The recent genomic sequencing of the marine bacterium Silicibacter sp. strain TM1040 revealed five prophage-like elements in its genome. The genomes of these prophages (named prophages 1 to 5) are approximately 74, 30, 39, 36, and 15 kb long, respectively. To understand the function of these prophages, cultures of TM1040 were treated with mitomycin C to induce the production of viral particles. A significant increase in viral counts and a decrease in bacterial counts when treated with mitomycin C suggested that prophages were induced from TM1040. Transmission electron microscopy revealed one dominant type of siphovirus, while pulsed-field gel electrophoresis demonstrated two major DNA bands, equivalent to 35 and 75 kb, in the lysate. PCR amplification with primer sets specific to each prophage detected the presence of prophages 1, 3, and 4 in the viral lysate, suggesting that these prophages are inducible, but not necessarily to the same level, while prophages 2 and 5 are likely defective or non-mitomycin C-inducible phages. The combination of traditional phage assays and modern microbial genomics provides a quick and efficient way to investigate the functions and inducibility of prophages, particularly for a host harboring multiple prophages with similar sizes and morphological features.
Figures


Similar articles
-
Lysogeny and sporulation in Bacillus isolates from the Gulf of Mexico.Appl Environ Microbiol. 2010 Feb;76(3):829-42. doi: 10.1128/AEM.01710-09. Epub 2009 Dec 11. Appl Environ Microbiol. 2010. PMID: 20008174 Free PMC article.
-
Characterization of Functional Prophages in Clostridium difficile.Methods Mol Biol. 2016;1476:143-65. doi: 10.1007/978-1-4939-6361-4_11. Methods Mol Biol. 2016. PMID: 27507339
-
Biological and genomic analysis of a PBSX-like defective phage induced from Bacillus pumilus AB94180.Arch Virol. 2014 Apr;159(4):739-52. doi: 10.1007/s00705-013-1898-x. Epub 2013 Oct 25. Arch Virol. 2014. PMID: 24154951
-
Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas?ISME J. 2008 Jun;2(6):579-89. doi: 10.1038/ismej.2008.35. ISME J. 2008. PMID: 18521076 Review.
-
Comparative genomics of phages and prophages in lactic acid bacteria.Antonie Van Leeuwenhoek. 2002 Aug;82(1-4):73-91. Antonie Van Leeuwenhoek. 2002. PMID: 12369206 Review.
Cited by
-
The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial 'mobilome'.Environ Microbiol. 2009 Nov;11(11):2935-51. doi: 10.1111/j.1462-2920.2009.02081.x. Epub 2009 Oct 14. Environ Microbiol. 2009. PMID: 19840100 Free PMC article.
-
Comparative genome analysis provides deep insights into Aeromonas hydrophila taxonomy and virulence-related factors.BMC Genomics. 2018 Sep 26;19(1):712. doi: 10.1186/s12864-018-5100-4. BMC Genomics. 2018. PMID: 30257645 Free PMC article.
-
Increased sensitivity of Aggregatibacter actinomycetemcomitans to human serum is mediated by induction of a bacteriophage.Mol Oral Microbiol. 2023 Feb;38(1):58-70. doi: 10.1111/omi.12378. Epub 2022 Jul 25. Mol Oral Microbiol. 2023. PMID: 35833243 Free PMC article.
-
Impact of Viral Lysis on the Composition of Bacterial Communities and Dissolved Organic Matter in Deep-Sea Sediments.Viruses. 2020 Aug 22;12(9):922. doi: 10.3390/v12090922. Viruses. 2020. PMID: 32842650 Free PMC article.
-
Genome sequences of two novel phages infecting marine roseobacters.Environ Microbiol. 2009 Aug;11(8):2055-64. doi: 10.1111/j.1462-2920.2009.01927.x. Environ Microbiol. 2009. PMID: 19689706 Free PMC article.
References
-
- Ackermann, H. W., and M. S. DuBow. 1987. Viruses of prokaryotes: general properties of bacteriophages. CRC Press, Inc., Boca Raton, Fla.
-
- Arber, W., L. Enquist, B. Hohn, N. E. Murray, and K. Murray. 1983. Experimental methods for use with lambda, p. 433-466. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
-
- Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. - PubMed
-
- Bordenstein, S. R., and W. S. Reznikoff. 2005. Mobile DNA in obligate intracellular bacteria. Nat. Rev. Microbiol. 3:688-698. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

